Tabular data¶
Alongside the timeseries data produced continuously at the laboratories, the gravitational-wave community produces a number of different sets of tabular data, including segments of time indicating interferometer state, and transient event triggers.
The LIGO_LW XML format¶
The LIGO Scientific Collaboration uses a custom scheme of XML in which to
store tabular data, called the LIGO_LW scheme.
Complementing the scheme is a python library - glue.ligolw - which
allows users to read and write all of the different types of tabular data
produced by gravitational-wave searches.
The remainder of this document outlines the small number of extensions that
GWpy provides for the table classes provided by glue.ligolw.
Note
All users should review the original documentation for glue.ligolw
to get any full sense of how to use these objects.
Reading LIGO_LW files¶
GWpy annotates all of the Table subclasses defined
in glue.ligolw.lsctables to make reading those tables from LIGO_LW
XML files a bit easier.
These annotations hook into the unified input/output scheme used for all of the other core data classes. For example, you can read a table of single-interferometer burst events as follows:
>>> from gwpy.table.lsctables import SnglBurstTable
>>> events = SnglBurstTable.read('../../gwpy/tests/data/H1-LDAS_STRAIN-968654552-10.xml.gz')
For full details, check out the read() documentation.
Plotting event triggers¶
The other annotation GWpy defines provides a simple plotting method for a number of classes.
We can extend the above example to include plotting:
>>> plot = events.plot('time', 'central_freq', color='snr', edgecolor='none', epoch=968654552)
>>> plot.set_xlim(968654552, 968654552+10)
>>> plot.set_ylabel('Frequency [Hz]')
>>> plot.set_yscale('log')
>>> plot.set_title('LIGO Hanford Observatory event triggers for GW100916')
>>> plot.add_colorbar(clim=[1, 5], label='Signal-to-noise ratio', cmap='hot_r')
>>> plot.show()
(Source code, png)
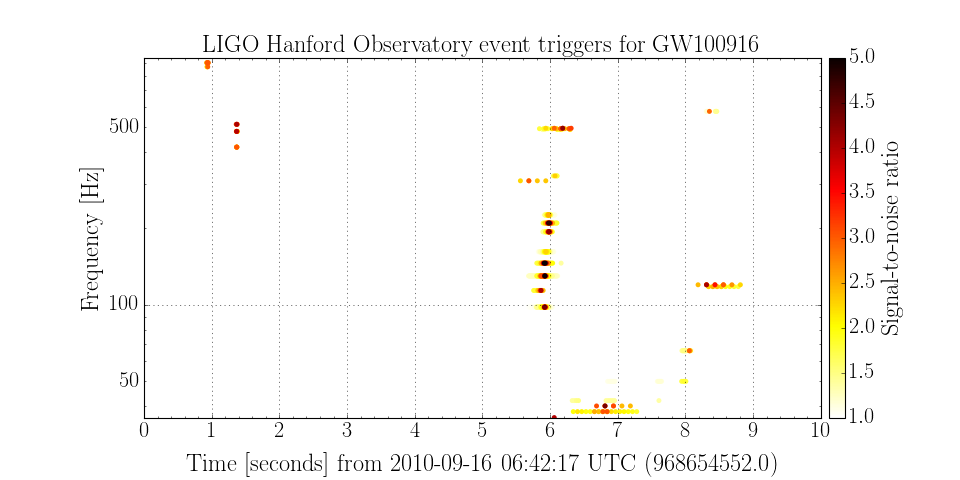
These code snippets are part of the GWpy example on plotting event triggers.
Plotting event tiles¶
Many types of event triggers define a 2-dimensional tile, for example in time and frequency. These tiles can be plotted in a similar manner to simple triggers.
>>> plot = events.plot('time', 'central_freq', 'duration', 'bandwidth', color='snr', epoch=968654552)
>>> plot.set_xlim(968654552, 968654552+10)
>>> plot.set_ylabel('Frequency [Hz]')
>>> plot.set_yscale('log')
>>> plot.set_title('LIGO Hanford Observatory event triggers for GW100916')
>>> plot.add_colorbar(clim=[1, 5], label='Signal-to-noise ratio', cmap='hot_r')
>>> plot.show()
(Source code, png)
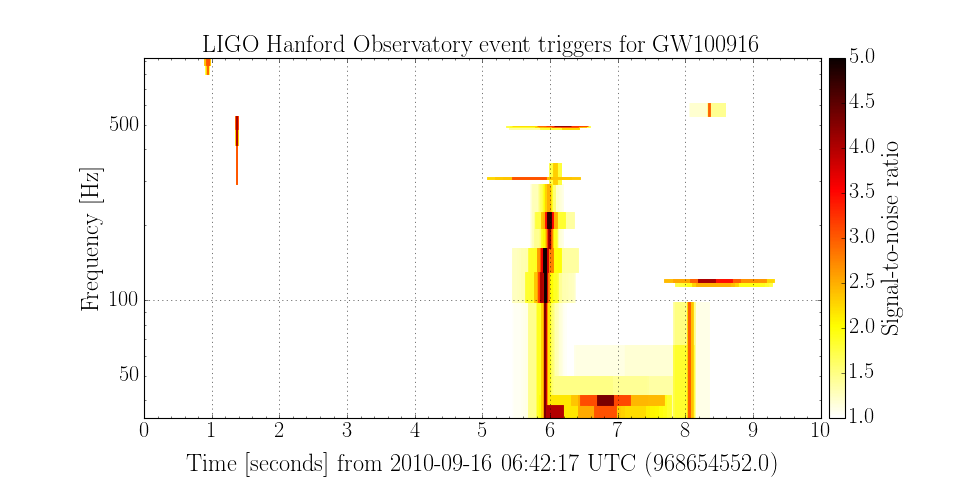
These code snippets are part of the GWpy example on plotting events as 2-d tiles.
Table applications¶
Class reference¶
Note
All of the below classes are based on glue.ligolw.table.Table.
This reference includes the following class entries:
-
class
gwpy.table.lsctables.CoincDefTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintshow_to_indexnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayget_coinc_def_id(search, search_coinc_type)Return the coinc_def_id for the row in the table whose search string and search_coinc_type integer have the values given. read(*args, **kwargs)Read data into a CoincDefTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (coinc_def_id)'¶
-
how_to_index= {'cd_ssct_index': ('search', 'search_coinc_type')}¶
-
next_id= <glue.ligolw.ilwd.coinc_definer_coinc_def_id_class object>¶
-
tableName= 'coinc_definer:table'¶
-
validcolumns= {'search': 'lstring', 'description': 'lstring', 'coinc_def_id': 'ilwd:char', 'search_coinc_type': 'int_4u'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_coinc_def_id(search, search_coinc_type, create_new=True, description=None)¶ Return the coinc_def_id for the row in the table whose search string and search_coinc_type integer have the values given. If a matching row is not found, the default behaviour is to create a new row and return the ID assigned to the new row. If, instead, create_new is False then KeyError is raised when a matching row is not found. The optional description parameter can be used to set the description string assigned to the new row if one is created, otherwise the new row is left with no description.
-
classmethod
read(*args, **kwargs)¶ Read data into a
CoincDefTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
CoincDefTableCoincDefTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes coinc_definer Yes No No csv Yes No No ligolw Yes No Yes
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.CoincTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintshow_to_indexinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayread(*args, **kwargs)Read data into a CoincTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (coinc_event_id)'¶
-
how_to_index= {'ce_tsi_index': ('time_slide_id',), 'ce_cdi_index': ('coinc_def_id',)}¶
-
interncolumns= ('process_id', 'coinc_def_id', 'time_slide_id', 'instruments')¶
-
next_id= <glue.ligolw.ilwd.coinc_event_coinc_event_id_class object>¶
-
tableName= 'coinc_event:table'¶
-
validcolumns= {'coinc_event_id': 'ilwd:char', 'instruments': 'lstring', 'nevents': 'int_4u', 'process_id': 'ilwd:char', 'coinc_def_id': 'ilwd:char', 'time_slide_id': 'ilwd:char', 'likelihood': 'real_8'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
classmethod
read(*args, **kwargs)¶ Read data into a
CoincTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
CoincTableCoincTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes coinc_event Yes No No csv Yes No No ligolw Yes No Yes
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.CoincInspiralTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
how_to_indexinterncolumnstableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a CoincInspiralTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
how_to_index= {'ci_cei_index': ('coinc_event_id',)}¶
-
interncolumns= ('coinc_event_id', 'ifos')¶
-
tableName= 'coinc_inspiral:table'¶
-
validcolumns= {'false_alarm_rate': 'real_8', 'mchirp': 'real_8', 'minimum_duration': 'real_8', 'mass': 'real_8', 'end_time': 'int_4s', 'coinc_event_id': 'ilwd:char', 'snr': 'real_8', 'end_time_ns': 'int_4s', 'combined_far': 'real_8', 'ifos': 'lstring'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
CoincInspiralTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
CoincInspiralTableCoincInspiralTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes coinc_inspiral Yes No No csv Yes No No ligolw Yes No Yes
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.CoincRingdownTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
how_to_indexinterncolumnstableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a CoincRingdownTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
how_to_index= {'cr_cei_index': ('coinc_event_id',)}¶
-
interncolumns= ('coinc_event_id', 'ifos')¶
-
tableName= 'coinc_ringdown:table'¶
-
validcolumns= {'snr_sq': 'real_8', 'false_alarm_rate': 'real_8', 'kappa': 'real_8', 'coinc_event_id': 'ilwd:char', 'spin': 'real_8', 'start_time': 'int_4s', 'start_time_ns': 'int_4s', 'combined_far': 'real_8', 'frequency': 'real_8', 'mass': 'real_8', 'choppedl_snr': 'real_8', 'snr': 'real_8', 'eff_coh_snr': 'real_8', 'quality': 'real_8', 'null_stat': 'real_8', 'snr_ratio': 'real_8', 'ifos': 'lstring'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
CoincRingdownTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
CoincRingdownTableCoincRingdownTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes coinc_ringdown Yes No No csv Yes No No ligolw Yes No Yes
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.ExperimentMapTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
how_to_indextableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayget_experiment_summ_ids(coinc_event_id)Gets all the experiment_summ_ids that map to a given coinc_event_id. read(*args, **kwargs)Read data into a ExperimentMapTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
how_to_index= {'em_cei_index': ('coinc_event_id',), 'em_esi_index': ('experiment_summ_id',)}¶
-
tableName= 'experiment_map:table'¶
-
validcolumns= {'experiment_summ_id': 'ilwd:char', 'coinc_event_id': 'ilwd:char'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_experiment_summ_ids(coinc_event_id)¶ Gets all the experiment_summ_ids that map to a given coinc_event_id.
-
classmethod
read(*args, **kwargs)¶ Read data into a
ExperimentMapTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
ExperimentMapTableExperimentMapTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No experiment_map Yes No No ligolw Yes No Yes
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.ExperimentSummaryTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsdatatypeshow_to_indexnext_idtableNamevalidcolumnsMethods Summary
add_nevents(experiment_summ_id, num_events)Add num_events to the nevents column in a specific entry in the table. as_id_dict()Return table as a dictionary mapping experiment_id, time_slide_id, veto_def_name, and sim_proc_id (if it exists) to the expr_summ_id. from_recarray(array[, columns])Create a new table from a numpy.recarrayget_expr_summ_id(experiment_id, ...[, ...])Return the expr_summ_id for the row in the table whose experiment_id, time_slide_id, veto_def_name, and datatype match the given. read(*args, **kwargs)Read data into a ExperimentSummaryTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarraywrite_experiment_summ(experiment_id, ...[, ...])Writes a single entry to the experiment_summ table. write_non_injection_summary(experiment_id, ...)Method for writing a new set of non-injection experiments to the experiment summary table. Attributes Documentation
-
constraints= 'PRIMARY KEY (experiment_summ_id)'¶
-
datatypes= ['slide', 'all_data', 'playground', 'exclude_play', 'simulation']¶
-
how_to_index= {'es_dt_index': ('datatype',), 'es_ei_index': ('experiment_id',)}¶
-
next_id= <glue.ligolw.ilwd.experiment_summary_experiment_summ_id_class object>¶
-
tableName= 'experiment_summary:table'¶
-
validcolumns= {'experiment_summ_id': 'ilwd:char', 'duration': 'int_4s', 'nevents': 'int_4u', 'sim_proc_id': 'ilwd:char', 'experiment_id': 'ilwd:char', 'datatype': 'lstring', 'time_slide_id': 'ilwd:char', 'veto_def_name': 'lstring'}¶
Methods Documentation
-
add_nevents(experiment_summ_id, num_events, add_to_current=True)¶ Add num_events to the nevents column in a specific entry in the table. If add_to_current is set to False, will overwrite the current nevents entry in the row with num_events. Otherwise, default is to add num_events to the current value.
Note: Can subtract events by passing a negative number to num_events.
-
as_id_dict()¶ Return table as a dictionary mapping experiment_id, time_slide_id, veto_def_name, and sim_proc_id (if it exists) to the expr_summ_id.
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_expr_summ_id(experiment_id, time_slide_id, veto_def_name, datatype, sim_proc_id=None)¶ Return the expr_summ_id for the row in the table whose experiment_id, time_slide_id, veto_def_name, and datatype match the given. If sim_proc_id, will retrieve the injection run matching that sim_proc_id. If a matching row is not found, returns None.
-
classmethod
read(*args, **kwargs)¶ Read data into a
ExperimentSummaryTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
ExperimentSummaryTableExperimentSummaryTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No experiment_summary Yes No No ligolw Yes No Yes
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
write_experiment_summ(experiment_id, time_slide_id, veto_def_name, datatype, sim_proc_id=None)¶ Writes a single entry to the experiment_summ table. This can be used for either injections or non-injection experiments. However, it is recommended that this only be used for injection experiments; for non-injection experiments write_experiment_summ_set should be used to ensure that an entry gets written for every time-slide performed.
-
write_non_injection_summary(experiment_id, time_slide_dict, veto_def_name, write_all_data=True, write_playground=True, write_exclude_play=True, return_dict=False)¶ Method for writing a new set of non-injection experiments to the experiment summary table. This ensures that for every entry in the experiment table, an entry for every slide is added to the experiment_summ table, rather than just an entry for slides that have events in them. Default is to write a 3 rows for zero-lag: one for all_data, playground, and exclude_play. (If all of these are set to false, will only slide rows.)
Note: sim_proc_id is hard-coded to None because time-slides are not performed with injections.
@experiment_id: the experiment_id for this experiment_summary set @time_slide_dict: the time_slide table as a dictionary; used to figure out
what is zero-lag and what is slide@veto_def_name: the name of the vetoes applied @write_all_data: if set to True, writes a zero-lag row who’s datatype column
is set to ‘all_data’@write_playground: same, but datatype is ‘playground’ @write_exclude_play: same, but datatype is ‘exclude_play’ @return_dict: if set to true, returns an id_dict of the table
-
-
class
gwpy.table.lsctables.ExperimentTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayget_expr_id(search_group, search, lars_id, ...)Return the expr_def_id for the row in the table whose values match the givens. get_row_from_id(experiment_id)Returns row in matching the given experiment_id. read(*args, **kwargs)Read data into a ExperimentTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarraywrite_new_expr_id(search_group, search, ...)Creates a new def_id for the given arguments and returns it. Attributes Documentation
-
constraints= 'PRIMARY KEY (experiment_id)'¶
-
next_id= <glue.ligolw.ilwd.experiment_experiment_id_class object>¶
-
tableName= 'experiment:table'¶
-
validcolumns= {'search': 'lstring', 'lars_id': 'lstring', 'experiment_id': 'ilwd:char', 'gps_start_time': 'int_4s', 'instruments': 'lstring', 'gps_end_time': 'int_4s', 'search_group': 'lstring', 'comments': 'lstring'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_expr_id(search_group, search, lars_id, instruments, gps_start_time, gps_end_time, comments=None)¶ Return the expr_def_id for the row in the table whose values match the givens. If a matching row is not found, returns None.
@search_group: string representing the search group (e.g., cbc) @serach: string representing search (e.g., inspiral) @lars_id: string representing lars_id @instruments: the instruments; must be a python set @gps_start_time: string or int representing the gps_start_time of the experiment @gps_end_time: string or int representing the gps_end_time of the experiment
-
get_row_from_id(experiment_id)¶ Returns row in matching the given experiment_id.
-
classmethod
read(*args, **kwargs)¶ Read data into a
ExperimentTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
ExperimentTableExperimentTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No experiment Yes No No ligolw Yes No Yes
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
write_new_expr_id(search_group, search, lars_id, instruments, gps_start_time, gps_end_time, comments=None)¶ Creates a new def_id for the given arguments and returns it. If an entry already exists with these, will just return that id.
@search_group: string representing the search group (e.g., cbc) @serach: string representing search (e.g., inspiral) @lars_id: string representing lars_id @instruments: the instruments; must be a python set @gps_start_time: string or int representing the gps_start_time of the experiment @gps_end_time: string or int representing the gps_end_time of the experiment
-
-
class
gwpy.table.lsctables.FilterTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a FilterTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (filter_id)'¶
-
next_id= <glue.ligolw.ilwd.filter_filter_id_class object>¶
-
tableName= 'filter:table'¶
-
validcolumns= {'comment': 'lstring', 'process_id': 'ilwd:char', 'param_set': 'int_4s', 'start_time': 'int_4s', 'filter_name': 'lstring', 'program': 'lstring', 'filter_id': 'ilwd:char'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
FilterTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
FilterTableFilterTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No filter Yes No No ligolw Yes No Yes
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.GDSTriggerTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a GDSTriggerTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (event_id)'¶
-
interncolumns= ('process_id', 'ifo', 'subtype')¶
-
next_id= <glue.ligolw.ilwd.gds_trigger_event_id_class object>¶
-
tableName= 'gds_trigger:table'¶
-
validcolumns= {'creator_db': 'int_4s', 'bandwidth': 'real_4', 'noise_power': 'real_4', 'duration': 'real_4', 'freq_peak': 'real_4', 'time_peak': 'real_4', 'freq_sigma': 'real_4', 'size': 'real_4', 'confidence': 'real_4', 'event_id': 'ilwd:char', 'priority': 'int_4s', 'significance': 'real_4', 'pixel_count': 'int_4s', 'start_time': 'int_4s', 'filter_id': 'ilwd:char', 'binarydata_length': 'int_4s', 'process_id': 'ilwd:char_u', 'disposition': 'int_4s', 'name': 'lstring', 'start_time_ns': 'int_4s', 'freq_average': 'real_4', 'time_average': 'real_4', 'binarydata': 'ilwd:char_u', 'subtype': 'lstring', 'frequency': 'real_4', 'time_sigma': 'real_4', 'ifo': 'lstring', 'signal_power': 'real_4'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
GDSTriggerTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
GDSTriggerTableGDSTriggerTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No gds_trigger Yes No No ligolw Yes No Yes
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.MultiBurstTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
how_to_indextableNamevalidcolumnsMethods Summary
binned_event_rates(stride, column, bins[, ...])Calculate an event rate TimeSeriesDictover a number of bins.event_rate(stride[, start, end, timecolumn])Calculate the rate TimeSeriesfor thisTable.from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a MultiBurstTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
how_to_index= {'mb_cei_index': ('coinc_event_id',)}¶
-
tableName= 'multi_burst:table'¶
-
validcolumns= {'central_freq': 'real_4', 'false_alarm_rate': 'real_4', 'confidence': 'real_4', 'creator_db': 'int_4s', 'ligo_angle_sig': 'real_4', 'coinc_event_id': 'ilwd:char', 'start_time_ns': 'int_4s', 'start_time': 'int_4s', 'ligo_axis_ra': 'real_4', 'bandwidth': 'real_4', 'process_id': 'ilwd:char', 'snr': 'real_4', 'ligo_angle': 'real_4', 'amplitude': 'real_4', 'filter_id': 'ilwd:char', 'duration': 'real_4', 'ligo_axis_dec': 'real_4', 'peak_time_ns': 'int_4s', 'peak_time': 'int_4s', 'ifos': 'lstring'}¶
Methods Documentation
-
binned_event_rates(stride, column, bins, operator='>=', start=None, end=None, timecolumn='time')[source]¶ Calculate an event rate
TimeSeriesDictover a number of bins.Parameters: stride :
floatsize (seconds) of each time bin
column :
strname of column by which to bin.
bins :
listone of:
'<','<=','>','>=','==','!=', for a standard mathematical operation,'in'to use the list of bins as containing bin edges, or- a callable function that takes compares an event value against the bin value and returns a boolean.
Note
If
binsis given as a list of tuples, this argument is ignored.start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeries.end :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rates :
TimeSeriesDicta dict of (bin,
TimeSeries) pairs describing a rate of events per second (Hz) for each of the bins.
-
event_rate(stride, start=None, end=None, timecolumn='time')[source]¶ Calculate the rate
TimeSeriesfor thisTable.Parameters: stride :
floatsize (seconds) of each time bin
start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeriesend :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rate :
TimeSeriesa
TimeSeriesof events per second (Hz)
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
MultiBurstTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
MultiBurstTableMultiBurstTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes multi_burst Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.MultiInspiralTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinstrument_idinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
binned_event_rates(stride, column, bins[, ...])Calculate an event rate TimeSeriesDictover a number of bins.event_rate(stride[, start, end, timecolumn])Calculate the rate TimeSeriesfor thisTable.from_recarray(array[, columns])Create a new table from a numpy.recarrayget_bestnr([index, nhigh, ...])Get the BestNR statistic for each row in the table get_coinc_chisq()@returns the coincident chisq for each row in the table get_coinc_snr()get_column(column)get_end()get_new_snr([index, nhigh, column])get_null_chisq()@returns the coherent null chisq for each row in the table get_null_snr()Get the coherent Null SNR for each row in the table. get_reduced_bank_chisq()@returns the bank chisq per degree of freedom for each row in get_reduced_chisq()@returns the chisq per degree of freedom for each row in get_reduced_coinc_chisq()@returns the coincident chisq per degree of freedom for each get_reduced_cont_chisq()@returns the auto (continuous) chisq per degree of freedom get_reduced_sngl_chisq(instrument)@returns the single-detector chisq per degree of freedom for get_sigmasq(instrument)Get the single-detector SNR of the given instrument for each row in the table. get_sigmasqs([instruments])Return dictionary of single-detector sigmas for each row in the table. get_sngl_bank_chisq(instrument)Get the single-detector chi^2 of the given instrument for each row in the table. get_sngl_bank_chisqs([instruments])Get the single-detector chi^2 for each row in the table. get_sngl_chisq(instrument)Get the single-detector chi^2 of the given instrument for each row in the table. get_sngl_chisqs([instruments])Get the single-detector chi^2 for each row in the table. get_sngl_cont_chisq(instrument)Get the single-detector chi^2 of the given instrument for each row in the table. get_sngl_cont_chisqs([instruments])Get the single-detector chi^2 for each row in the table. get_sngl_new_snr(ifo[, column, index, nhigh])get_sngl_snr(instrument)Get the single-detector SNR of the given instrument for each row in the table. get_sngl_snrs([instruments])Get the single-detector SNRs for each row in the table. getslide(slide_num)Return the triggers with a specific slide number. getstat()hist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a MultiInspiralTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayveto(seglist)vetoed(seglist)Return the inverse of what veto returns, i.e., return the triggers that lie within a given seglist. Attributes Documentation
-
constraints= 'PRIMARY KEY (event_id)'¶
-
instrument_id= {'G1': 'g', 'H2': 'h2', 'H1': 'h1', 'T1': 't', 'V1': 'v', 'L1': 'l'}¶
-
interncolumns= ('process_id', 'ifos', 'search')¶
-
next_id= <glue.ligolw.ilwd.multi_inspiral_event_id_class object>¶
-
tableName= 'multi_inspiral:table'¶
-
validcolumns= {'amp_term_1': 'real_4', 'amp_term_2': 'real_4', 'amp_term_3': 'real_4', 'amp_term_4': 'real_4', 'amp_term_5': 'real_4', 'amp_term_6': 'real_4', 'amp_term_7': 'real_4', 'amp_term_8': 'real_4', 'amp_term_9': 'real_4', 'chisq_dof': 'int_4s', 't1quad_re': 'real_4', 'autoCorrCohSq': 'real_4', 'v1quad_im': 'real_4', 'sigmasq_h2': 'real_8', 'null_stat_h1h2': 'real_4', 'sigmasq_h1': 'real_8', 'bank_chisq_t': 'real_4', 'bank_chisq_v': 'real_4', 'bank_chisq_l': 'real_4', 'sngl_bank_chisq_dof': 'int_4s', 'ttotal': 'real_4', 'bank_chisq_g': 'real_4', 'snr_h1': 'real_4', 'snr_h2': 'real_4', 'h2quad_re': 'real_4', 'process_id': 'ilwd:char', 'sngl_chisq_dof': 'int_4s', 'end_time': 'int_4s', 'end_time_ns': 'int_4s', 'eff_dist_h1h2': 'real_4', 'h2quad_im': 'real_4', 'ligo_angle_sig': 'real_4', 'cont_chisq_h1': 'real_4', 'l1quad_im': 'real_4', 'ra': 'real_4', 'impulse_time_ns': 'int_4s', 'inclination': 'real_4', 't1quad_im': 'real_4', 'bank_chisq_dof': 'int_4s', 'amp_term_10': 'real_4', 'cont_chisq_g': 'real_4', 'search': 'lstring', 'cont_chisq_l': 'real_4', 'cont_chisq_v': 'real_4', 'l1quad_re': 'real_4', 'cont_chisq_t': 'real_4', 'eta': 'real_4', 'ligo_angle': 'real_4', 'bank_chisq': 'real_4', 'g1quad_re': 'real_4', 'end_time_gmst': 'real_8', 'snr_dof': 'int_4s', 'v1quad_re': 'real_4', 'cont_chisq_h2': 'real_4', 'coa_phase': 'real_4', 'ifos': 'lstring', 'coh_snr_h1h2': 'real_4', 'mchirp': 'real_4', 'tau3': 'real_4', 'chi': 'real_4', 'tau2': 'real_4', 'bank_chisq_h1': 'real_4', 'tau0': 'real_4', 'autoCorrNullSq': 'real_4', 'tau4': 'real_4', 'tau5': 'real_4', 'h1quad_im': 'real_4', 'impulse_time': 'int_4s', 'crossCorrCohSq': 'real_4', 'mass1': 'real_4', 'ampMetricEigenVal2': 'real_8', 'ampMetricEigenVal1': 'real_8', 'distance': 'real_4', 'g1quad_im': 'real_4', 'kappa': 'real_4', 'crossCorrNullSq': 'real_4', 'h1quad_re': 'real_4', 'sngl_cont_chisq_dof': 'int_4s', 'amplitude': 'real_4', 'cohSnrSqLocal': 'real_4', 'dec': 'real_4', 'eff_dist_l': 'real_4', 'chisq_h1': 'real_4', 'chisq_h2': 'real_4', 'eff_dist_g': 'real_4', 'bank_chisq_h2': 'real_4', 'chisq': 'real_4', 'eff_dist_v': 'real_4', 'eff_dist_t': 'real_4', 'sigmasq_t': 'real_8', 'sigmasq_v': 'real_8', 'null_statistic': 'real_4', 'snr_l': 'real_4', 'event_id': 'ilwd:char', 'snr_g': 'real_4', 'polarization': 'real_4', 'cont_chisq': 'real_4', 'sigmasq_g': 'real_8', 'null_stat_degen': 'real_4', 'sigmasq_l': 'real_8', 'time_slide_id': 'ilwd:char', 'snr_v': 'real_4', 'snr_t': 'real_4', 'cont_chisq_dof': 'int_4s', 'trace_snr': 'real_4', 'snr': 'real_4', 'eff_dist_h1': 'real_4', 'eff_dist_h2': 'real_4', 'chisq_t': 'real_4', 'chisq_v': 'real_4', 'mass2': 'real_4', 'chisq_g': 'real_4', 'chisq_l': 'real_4'}¶
Methods Documentation
-
binned_event_rates(stride, column, bins, operator='>=', start=None, end=None, timecolumn='time')[source]¶ Calculate an event rate
TimeSeriesDictover a number of bins.Parameters: stride :
floatsize (seconds) of each time bin
column :
strname of column by which to bin.
bins :
listone of:
'<','<=','>','>=','==','!=', for a standard mathematical operation,'in'to use the list of bins as containing bin edges, or- a callable function that takes compares an event value against the bin value and returns a boolean.
Note
If
binsis given as a list of tuples, this argument is ignored.start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeries.end :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rates :
TimeSeriesDicta dict of (bin,
TimeSeries) pairs describing a rate of events per second (Hz) for each of the bins.
-
event_rate(stride, start=None, end=None, timecolumn='time')[source]¶ Calculate the rate
TimeSeriesfor thisTable.Parameters: stride :
floatsize (seconds) of each time bin
start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeriesend :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rate :
TimeSeriesa
TimeSeriesof events per second (Hz)
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_bestnr(index=4.0, nhigh=3.0, null_snr_threshold=4.25, null_grad_thresh=20.0, null_grad_val=0.2)¶ Get the BestNR statistic for each row in the table
-
get_coinc_chisq()¶ @returns the coincident chisq for each row in the table
-
get_coinc_snr()¶
-
get_column(column)¶
-
get_end()¶
-
get_new_snr(index=4.0, nhigh=3.0, column='chisq')¶
-
get_null_chisq()¶ @returns the coherent null chisq for each row in the table
-
get_null_snr()¶ Get the coherent Null SNR for each row in the table.
-
get_reduced_bank_chisq()¶ @returns the bank chisq per degree of freedom for each row in this table
-
get_reduced_chisq()¶ @returns the chisq per degree of freedom for each row in this table
-
get_reduced_coinc_chisq()¶ @returns the coincident chisq per degree of freedom for each row in the table
-
get_reduced_cont_chisq()¶ @returns the auto (continuous) chisq per degree of freedom for each row in this table
-
get_reduced_sngl_chisq(instrument)¶ @returns the single-detector chisq per degree of freedom for each row in this table
-
get_sigmasq(instrument)¶ Get the single-detector SNR of the given instrument for each row in the table.
-
get_sigmasqs(instruments=None)¶ Return dictionary of single-detector sigmas for each row in the table.
-
get_sngl_bank_chisq(instrument)¶ Get the single-detector chi^2 of the given instrument for each row in the table.
-
get_sngl_bank_chisqs(instruments=None)¶ Get the single-detector chi^2 for each row in the table.
-
get_sngl_chisq(instrument)¶ Get the single-detector chi^2 of the given instrument for each row in the table.
-
get_sngl_chisqs(instruments=None)¶ Get the single-detector chi^2 for each row in the table.
-
get_sngl_cont_chisq(instrument)¶ Get the single-detector chi^2 of the given instrument for each row in the table.
-
get_sngl_cont_chisqs(instruments=None)¶ Get the single-detector chi^2 for each row in the table.
-
get_sngl_new_snr(ifo, column='chisq', index=4.0, nhigh=3.0)¶
-
get_sngl_snr(instrument)¶ Get the single-detector SNR of the given instrument for each row in the table.
-
get_sngl_snrs(instruments=None)¶ Get the single-detector SNRs for each row in the table.
-
getslide(slide_num)¶ Return the triggers with a specific slide number. @param slide_num: the slide number to recover (contained in the event_id)
-
getstat()¶
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
MultiInspiralTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
MultiInspiralTableMultiInspiralTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes multi_inspiral Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
veto(seglist)¶
-
vetoed(seglist)¶ Return the inverse of what veto returns, i.e., return the triggers that lie within a given seglist.
-
-
class
gwpy.table.lsctables.ProcessTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayget_ids_by_program(program)Return a set containing the process IDs from rows whose program string equals the given program. hist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a ProcessTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (process_id)'¶
-
next_id= <glue.ligolw.ilwd.process_process_id_class object>¶
-
tableName= 'process:table'¶
-
validcolumns= {'comment': 'lstring', 'node': 'lstring', 'domain': 'lstring', 'unix_procid': 'int_4s', 'start_time': 'int_4s', 'process_id': 'ilwd:char', 'is_online': 'int_4s', 'ifos': 'lstring', 'jobid': 'int_4s', 'username': 'lstring', 'program': 'lstring', 'end_time': 'int_4s', 'version': 'lstring', 'cvs_repository': 'lstring', 'cvs_entry_time': 'int_4s'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_ids_by_program(program)¶ Return a set containing the process IDs from rows whose program string equals the given program.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
ProcessTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
ProcessTableProcessTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes process Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.ProcessParamsTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
how_to_indextableNamevalidcolumnsMethods Summary
append(row)from_recarray(array[, columns])Create a new table from a numpy.recarrayread(*args, **kwargs)Read data into a ProcessParamsTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
how_to_index= {'pp_pip_index': ('process_id', 'param')}¶
-
tableName= 'process_params:table'¶
-
validcolumns= {'process_id': 'ilwd:char', 'program': 'lstring', 'type': 'lstring', 'value': 'lstring', 'param': 'lstring'}¶
Methods Documentation
-
append(row)¶
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
classmethod
read(*args, **kwargs)¶ Read data into a
ProcessParamsTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
ProcessParamsTableProcessParamsTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes process_params Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SearchSummaryTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
how_to_indextableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayget_in_segmentlistdict([process_ids])Return a segmentlistdict mapping instrument to in segment list. get_inlist()Return a segmentlist object describing the times spanned by the input segments of all rows in the table. get_out_segmentlistdict([process_ids])Return a segmentlistdict mapping instrument to out segment list. get_outlist()Return a segmentlist object describing the times spanned by the output segments of all rows in the table. read(*args, **kwargs)Read data into a SearchSummaryTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
how_to_index= {'ss_pi_index': ('process_id',)}¶
-
tableName= 'search_summary:table'¶
-
validcolumns= {'comment': 'lstring', 'nevents': 'int_4s', 'lal_cvs_tag': 'lstring', 'out_start_time': 'int_4s', 'shared_object': 'lstring', 'in_start_time': 'int_4s', 'nnodes': 'int_4s', 'out_start_time_ns': 'int_4s', 'process_id': 'ilwd:char', 'in_end_time': 'int_4s', 'out_end_time_ns': 'int_4s', 'lalwrapper_cvs_tag': 'lstring', 'in_start_time_ns': 'int_4s', 'in_end_time_ns': 'int_4s', 'out_end_time': 'int_4s', 'ifos': 'lstring'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_in_segmentlistdict(process_ids=None)¶ Return a segmentlistdict mapping instrument to in segment list. If process_ids is a sequence of process IDs, then only rows with matching IDs are included otherwise all rows are included.
Note: the result is not coalesced, each segmentlist contains the segments listed for that instrument as they appeared in the table.
-
get_inlist()¶ Return a segmentlist object describing the times spanned by the input segments of all rows in the table.
Note: the result is not coalesced, the segmentlist contains the segments as they appear in the table.
-
get_out_segmentlistdict(process_ids=None)¶ Return a segmentlistdict mapping instrument to out segment list. If process_ids is a sequence of process IDs, then only rows with matching IDs are included otherwise all rows are included.
Note: the result is not coalesced, each segmentlist contains the segments listed for that instrument as they appeared in the table.
-
get_outlist()¶ Return a segmentlist object describing the times spanned by the output segments of all rows in the table.
Note: the result is not coalesced, the segmentlist contains the segments as they appear in the table.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SearchSummaryTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SearchSummaryTableSearchSummaryTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes search_summary Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SearchSummVarsTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayread(*args, **kwargs)Read data into a SearchSummVarsTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (search_summvar_id)'¶
-
next_id= <glue.ligolw.ilwd.search_summvars_search_summvar_id_class object>¶
-
tableName= 'search_summvars:table'¶
-
validcolumns= {'search_summvar_id': 'ilwd:char', 'process_id': 'ilwd:char', 'name': 'lstring', 'value': 'real_8', 'string': 'lstring'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SearchSummVarsTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SearchSummVarsTableSearchSummVarsTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes search_summvars Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SimBurstTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayread(*args, **kwargs)Read data into a SimBurstTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (simulation_id)'¶
-
interncolumns= ('process_id', 'waveform')¶
-
next_id= <glue.ligolw.ilwd.sim_burst_simulation_id_class object>¶
-
tableName= 'sim_burst:table'¶
-
validcolumns= {'hrss': 'real_8', 'time_geocent_gps': 'int_4s', 'psi': 'real_8', 'amplitude': 'real_8', 'egw_over_rsquared': 'real_8', 'waveform_number': 'int_8u', 'pol_ellipse_angle': 'real_8', 'simulation_id': 'ilwd:char', 'q': 'real_8', 'waveform': 'lstring', 'bandwidth': 'real_8', 'process_id': 'ilwd:char', 'frequency': 'real_8', 'ra': 'real_8', 'time_geocent_gmst': 'real_8', 'pol_ellipse_e': 'real_8', 'duration': 'real_8', 'time_slide_id': 'ilwd:char', 'dec': 'real_8', 'time_geocent_gps_ns': 'int_4s'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SimBurstTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SimBurstTableSimBurstTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes sim_burst Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SimInspiralTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayget_chirp_dist([ref_mass])get_chirp_eff_dist(site[, ref_mass])get_column(column)get_end([site])get_spin_mag(objectnumber)read(*args, **kwargs)Read data into a SimInspiralTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayveto(seglist)vetoed(seglist)Return the inverse of what veto returns, i.e., return the triggers that lie within a given seglist. Attributes Documentation
-
constraints= 'PRIMARY KEY (simulation_id)'¶
-
interncolumns= ('process_id', 'waveform', 'source')¶
-
next_id= <glue.ligolw.ilwd.sim_inspiral_simulation_id_class object>¶
-
tableName= 'sim_inspiral:table'¶
-
validcolumns= {'theta0': 'real_4', 'geocent_end_time_ns': 'int_4s', 'amp_order': 'int_4s', 'end_time_gmst': 'real_8', 'coa_phase': 'real_4', 'mchirp': 'real_4', 'numrel_mode_min': 'int_4s', 'numrel_mode_max': 'int_4s', 'source': 'lstring', 'latitude': 'real_4', 'numrel_data': 'lstring', 'geocent_end_time': 'int_4s', 'spin2x': 'real_4', 'spin2y': 'real_4', 'spin2z': 'real_4', 'process_id': 'ilwd:char', 'h_end_time': 'int_4s', 'distance': 'real_4', 't_end_time': 'int_4s', 'taper': 'lstring', 'longitude': 'real_4', 'v_end_time_ns': 'int_4s', 'bandpass': 'int_4s', 'eff_dist_l': 'real_4', 'eff_dist_h': 'real_4', 'eff_dist_g': 'real_4', 't_end_time_ns': 'int_4s', 'spin1y': 'real_4', 'spin1x': 'real_4', 'spin1z': 'real_4', 'h_end_time_ns': 'int_4s', 'eff_dist_t': 'real_4', 'l_end_time_ns': 'int_4s', 'alpha2': 'real_4', 'alpha3': 'real_4', 'alpha1': 'real_4', 'alpha6': 'real_4', 'alpha4': 'real_4', 'alpha5': 'real_4', 'l_end_time': 'int_4s', 'polarization': 'real_4', 'waveform': 'lstring', 'phi0': 'real_4', 'inclination': 'real_4', 'simulation_id': 'ilwd:char', 'f_lower': 'real_4', 'g_end_time_ns': 'int_4s', 'eff_dist_v': 'real_4', 'beta': 'real_4', 'g_end_time': 'int_4s', 'alpha': 'real_4', 'f_final': 'real_4', 'mass1': 'real_4', 'mass2': 'real_4', 'v_end_time': 'int_4s', 'eta': 'real_4', 'psi0': 'real_4', 'psi3': 'real_4'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_chirp_dist(ref_mass=1.4)¶
-
get_chirp_eff_dist(site, ref_mass=1.4)¶
-
get_column(column)¶
-
get_end(site=None)¶
-
get_spin_mag(objectnumber)¶
-
classmethod
read(*args, **kwargs)¶ Read data into a
SimInspiralTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SimInspiralTableSimInspiralTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes sim_inspiral Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
veto(seglist)¶
-
vetoed(seglist)¶ Return the inverse of what veto returns, i.e., return the triggers that lie within a given seglist.
-
-
class
gwpy.table.lsctables.SimRingdownTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayread(*args, **kwargs)Read data into a SimRingdownTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (simulation_id)'¶
-
interncolumns= ('process_id', 'waveform', 'coordinates')¶
-
next_id= <glue.ligolw.ilwd.sim_ringdown_simulation_id_class object>¶
-
tableName= 'sim_ringdown:table'¶
-
validcolumns= {'simulation_id': 'ilwd:char', 'eff_dist_l': 'real_4', 'hrss': 'real_4', 'v_start_time_ns': 'int_4s', 'eff_dist_h': 'real_4', 'epsilon': 'real_4', 'process_id': 'ilwd:char', 'start_time_gmst': 'real_8', 'phase': 'real_4', 'eff_dist_v': 'real_4', 'spin': 'real_4', 'quality': 'real_4', 'h_start_time': 'int_4s', 'distance': 'real_4', 'hrss_l': 'real_4', 'hrss_h': 'real_4', 'h_start_time_ns': 'int_4s', 'v_start_time': 'int_4s', 'coordinates': 'lstring', 'polarization': 'real_4', 'waveform': 'lstring', 'frequency': 'real_4', 'mass': 'real_4', 'longitude': 'real_4', 'amplitude': 'real_4', 'geocent_start_time_ns': 'int_4s', 'latitude': 'real_4', 'hrss_v': 'real_4', 'geocent_start_time': 'int_4s', 'l_start_time': 'int_4s', 'l_start_time_ns': 'int_4s', 'inclination': 'real_4'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SimRingdownTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SimRingdownTableSimRingdownTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes sim_ringdown Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SnglBurstTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
binned_event_rates(stride, column, bins[, ...])Calculate an event rate TimeSeriesDictover a number of bins.event_rate(stride[, start, end, timecolumn])Calculate the rate TimeSeriesfor thisTable.fetch(channel, etg, start, end[, verbose])Find and read events into a SnglBurstTable.from_recarray(array[, columns])Create a new table from a numpy.recarrayget_band()@returns: the frequency band of each row in the table get_column(column)@returns: an array of column values for each row in the table get_ms_band()@returns: the frequency band of the most significant tile get_ms_period()@returns: the period segment for the most significant tile get_ms_start()@returns: the start time of the most significant tile for get_ms_stop()@returns: the stop time of the most significant tile for get_peak()@returns: the peak time of each row in the table get_period()@returns: the period segment of each row in the table get_q()@returns: the Q of each row in the table get_start()@returns: the start time of each row in the table get_stop()@returns: the stop time of each row in the table get_z()@returns: the Z (Omega-Pipeline energy) of each row in the hist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a SnglBurstTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayveto(seglist)@returns: those rows of the table that don’t lie within a veto_seglistdict(seglistdict)vetoed(seglist)@returns: those rows of the table that lie within a given vetoed_seglistdict(seglistdict)Attributes Documentation
-
constraints= 'PRIMARY KEY (event_id)'¶
-
interncolumns= ('process_id', 'ifo', 'search', 'channel')¶
-
next_id= <glue.ligolw.ilwd.sngl_burst_event_id_class object>¶
-
tableName= 'sngl_burst:table'¶
-
validcolumns= {'param_two_value': 'real_8', 'chisq_dof': 'real_8', 'bandwidth': 'real_4', 'peak_time_error': 'real_4', 'central_freq': 'real_4', 'confidence': 'real_4', 'peak_frequency': 'real_4', 'peak_strain_error': 'real_4', 'ms_snr': 'real_4', 'filter_id': 'ilwd:char', 'time_lag': 'real_4', 'peak_time_ns': 'int_4s', 'start_time': 'int_4s', 'process_id': 'ilwd:char', 'fhigh': 'real_4', 'ms_stop_time_ns': 'int_4s', 'ms_start_time_ns': 'int_4s', 'param_one_name': 'lstring', 'stop_time_ns': 'int_4s', 'channel': 'lstring', 'amplitude': 'real_4', 'ms_duration': 'real_4', 'ifo': 'lstring', 'creator_db': 'int_4s', 'peak_strain': 'real_4', 'param_two_name': 'lstring', 'chisq': 'real_8', 'ms_flow': 'real_4', 'duration': 'real_4', 'ms_start_time': 'int_4s', 'param_three_name': 'lstring', 'event_id': 'ilwd:char', 'peak_frequency_error': 'real_4', 'param_one_value': 'real_8', 'param_three_value': 'real_8', 'hrss': 'real_4', 'ms_hrss': 'real_4', 'stop_time': 'int_4s', 'peak_time': 'int_4s', 'ms_fhigh': 'real_4', 'ms_confidence': 'real_4', 'snr': 'real_4', 'search': 'lstring', 'start_time_ns': 'int_4s', 'tfvolume': 'real_4', 'flow': 'real_4', 'ms_bandwidth': 'real_4', 'ms_stop_time': 'int_4s'}¶
Methods Documentation
-
binned_event_rates(stride, column, bins, operator='>=', start=None, end=None, timecolumn='time')[source]¶ Calculate an event rate
TimeSeriesDictover a number of bins.Parameters: stride :
floatsize (seconds) of each time bin
column :
strname of column by which to bin.
bins :
listone of:
'<','<=','>','>=','==','!=', for a standard mathematical operation,'in'to use the list of bins as containing bin edges, or- a callable function that takes compares an event value against the bin value and returns a boolean.
Note
If
binsis given as a list of tuples, this argument is ignored.start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeries.end :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rates :
TimeSeriesDicta dict of (bin,
TimeSeries) pairs describing a rate of events per second (Hz) for each of the bins.
-
event_rate(stride, start=None, end=None, timecolumn='time')[source]¶ Calculate the rate
TimeSeriesfor thisTable.Parameters: stride :
floatsize (seconds) of each time bin
start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeriesend :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rate :
TimeSeriesa
TimeSeriesof events per second (Hz)
-
classmethod
fetch(channel, etg, start, end, verbose=False, **kwargs)[source]¶ Find and read events into a
SnglBurstTable.Event XML files are searched for only on the LIGO Data Grid using the conventions set out in LIGO-T1300468.
Warning
This method will not work on machines outside of the LIGO Lab computing centres at Caltech, LHO, and LHO.
Parameters: channel :
strthe name of the data channel to search for
etg :
strthe name of the event trigger generator (ETG)
start :
float,LIGOTimeGPSthe GPS start time of the search
end :
float,LIGOTimeGPSthe GPS end time of the search
verbose :
bool, optionalprint verbose output, default:
False**kwargs :
other keyword arguments to pass to
SnglBurstTable.read()Returns: table :
SnglBurstTablea new
SnglBurstTablecontaining triggers read from the standard pathsRaises: ValueError :
if no channel-level directory is found for the given channel, indicating that nothing has ever been processed for this channel.
See also
SnglBurstTable.read- for documentation of the available keyword arguments
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_band()¶ @returns: the frequency band of each row in the table @returntype: glue.segments.segmentlist
-
get_column(column)¶ @returns: an array of column values for each row in the table
- @param column:
- name of column to return
- @returntype:
- numpy.ndarray
-
get_ms_band()¶ @returns: the frequency band of the most significant tile for each row in the table
-
get_ms_period()¶ @returns: the period segment for the most significant tile of each row in the table @returntype: glue.segments.segmentlist
-
get_ms_start()¶ @returns: the start time of the most significant tile for each row in the table @returntype: numpy.ndarray
-
get_ms_stop()¶ @returns: the stop time of the most significant tile for each row in the table @returntype: numpy.ndarray
-
get_peak()¶ @returns: the peak time of each row in the table @returntype: numpy.ndarray
-
get_period()¶ @returns: the period segment of each row in the table @returntype: glue.segments.segmentlist
-
get_q()¶ @returns: the Q of each row in the table @returntype: numpy.ndarray
-
get_start()¶ @returns: the start time of each row in the table @returntype: numpy.ndarray
-
get_stop()¶ @returns: the stop time of each row in the table @returntype: numpy.ndarray
-
get_z()¶ @returns: the Z (Omega-Pipeline energy) of each row in the table @returntype: numpy.ndarray
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SnglBurstTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SnglBurstTableSnglBurstTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No Yes cwb Yes No No cwb-ascii Yes No Yes ligolw Yes No Yes omega Yes No No omegadq Yes No No omicron Yes No Yes sngl_burst Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
veto(seglist)¶ @returns: those rows of the table that don’t lie within a given seglist
-
veto_seglistdict(seglistdict)¶
-
vetoed(seglist)¶ @returns: those rows of the table that lie within a given seglist
-
vetoed_seglistdict(seglistdict)¶
-
-
class
gwpy.table.lsctables.SnglInspiralTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
binned_event_rates(stride, column, bins[, ...])Calculate an event rate TimeSeriesDictover a number of bins.event_rate(stride[, start, end, timecolumn])Calculate the rate TimeSeriesfor thisTable.from_recarray(array[, columns])Create a new table from a numpy.recarrayget_bank_effective_snr([fac])get_bank_new_snr([index])get_chirp_eff_dist([ref_mass])get_column(column[, fac, index])get_cont_effective_snr([fac])get_cont_new_snr([index])get_effective_snr([fac])get_end()get_lvS5stat()get_new_snr([index])get_reduced_bank_chisq()get_reduced_chisq()get_reduced_cont_chisq()get_snr_over_chi()getslide(slide_num)Return the triggers with a specific slide number. hist(column, **kwargs)Generate a HistogramPlotof thisTable.ifocut(ifo[, inplace])Return a SnglInspiralTable with rows from self having IFO equal to the given ifo. plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a SnglInspiralTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayveto(seglist)veto_seglistdict(seglistdict)vetoed(seglist)Return the inverse of what veto returns, i.e., return the triggers that lie within a given seglist. vetoed_seglistdict(seglistdict)Attributes Documentation
-
constraints= 'PRIMARY KEY (event_id)'¶
-
interncolumns= ('process_id', 'ifo', 'search', 'channel')¶
-
next_id= <glue.ligolw.ilwd.sngl_inspiral_event_id_class object>¶
-
tableName= 'sngl_inspiral:table'¶
-
validcolumns= {'cont_chisq': 'real_4', 'bank_chisq': 'real_4', 'chisq_dof': 'int_4s', 'end_time_gmst': 'real_8', 'event_duration': 'real_8', 'chisq': 'real_4', 'spin1y': 'real_4', 'spin1x': 'real_4', 'alpha': 'real_4', 'coa_phase': 'real_4', 'alpha2': 'real_4', 'mchirp': 'real_4', 'alpha1': 'real_4', 'alpha6': 'real_4', 'alpha4': 'real_4', 'alpha5': 'real_4', 'event_id': 'ilwd:char', 'chi': 'real_4', 'cont_chisq_dof': 'int_4s', 'spin2y': 'real_4', 'tau2': 'real_4', 'tau3': 'real_4', 'tau0': 'real_4', 'tau4': 'real_4', 'tau5': 'real_4', 'template_duration': 'real_8', 'impulse_time': 'int_4s', 'impulse_time_ns': 'int_4s', 'rsqveto_duration': 'real_4', 'channel': 'lstring', 'mtotal': 'real_4', 'alpha3': 'real_4', 'spin1z': 'real_4', 'Gamma5': 'real_4', 'spin2x': 'real_4', 'f_final': 'real_4', 'beta': 'real_4', 'process_id': 'ilwd:char', 'snr': 'real_4', 'bank_chisq_dof': 'int_4s', 'kappa': 'real_4', 'eff_distance': 'real_4', 'Gamma7': 'real_4', 'Gamma6': 'real_4', 'search': 'lstring', 'Gamma4': 'real_4', 'mass1': 'real_4', 'Gamma2': 'real_4', 'Gamma1': 'real_4', 'mass2': 'real_4', 'ttotal': 'real_4', 'Gamma0': 'real_4', 'spin2z': 'real_4', 'Gamma9': 'real_4', 'Gamma8': 'real_4', 'Gamma3': 'real_4', 'eta': 'real_4', 'psi0': 'real_4', 'end_time': 'int_4s', 'amplitude': 'real_4', 'psi3': 'real_4', 'end_time_ns': 'int_4s', 'ifo': 'lstring', 'sigmasq': 'real_8'}¶
Methods Documentation
-
binned_event_rates(stride, column, bins, operator='>=', start=None, end=None, timecolumn='time')[source]¶ Calculate an event rate
TimeSeriesDictover a number of bins.Parameters: stride :
floatsize (seconds) of each time bin
column :
strname of column by which to bin.
bins :
listone of:
'<','<=','>','>=','==','!=', for a standard mathematical operation,'in'to use the list of bins as containing bin edges, or- a callable function that takes compares an event value against the bin value and returns a boolean.
Note
If
binsis given as a list of tuples, this argument is ignored.start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeries.end :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rates :
TimeSeriesDicta dict of (bin,
TimeSeries) pairs describing a rate of events per second (Hz) for each of the bins.
-
event_rate(stride, start=None, end=None, timecolumn='time')[source]¶ Calculate the rate
TimeSeriesfor thisTable.Parameters: stride :
floatsize (seconds) of each time bin
start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeriesend :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rate :
TimeSeriesa
TimeSeriesof events per second (Hz)
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_bank_effective_snr(fac=250.0)¶
-
get_bank_new_snr(index=6.0)¶
-
get_chirp_eff_dist(ref_mass=1.4)¶
-
get_column(column, fac=250.0, index=6.0)¶
-
get_cont_effective_snr(fac=250.0)¶
-
get_cont_new_snr(index=6.0)¶
-
get_effective_snr(fac=250.0)¶
-
get_end()¶
-
get_lvS5stat()¶
-
get_new_snr(index=6.0)¶
-
get_reduced_bank_chisq()¶
-
get_reduced_chisq()¶
-
get_reduced_cont_chisq()¶
-
get_snr_over_chi()¶
-
getslide(slide_num)¶ Return the triggers with a specific slide number. @param slide_num: the slide number to recover (contained in the event_id)
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
ifocut(ifo, inplace=False)¶ Return a SnglInspiralTable with rows from self having IFO equal to the given ifo. If inplace, modify self directly, else create a new table and fill it.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SnglInspiralTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SnglInspiralTableSnglInspiralTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes sngl_inspiral Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
veto(seglist)¶
-
veto_seglistdict(seglistdict)¶
-
vetoed(seglist)¶ Return the inverse of what veto returns, i.e., return the triggers that lie within a given seglist.
-
vetoed_seglistdict(seglistdict)¶
-
-
class
gwpy.table.lsctables.SnglRingdownTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
binned_event_rates(stride, column, bins[, ...])Calculate an event rate TimeSeriesDictover a number of bins.event_rate(stride[, start, end, timecolumn])Calculate the rate TimeSeriesfor thisTable.from_recarray(array[, columns])Create a new table from a numpy.recarrayget_start()hist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a SnglRingdownTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (event_id)'¶
-
interncolumns= ('process_id', 'ifo', 'channel')¶
-
next_id= <glue.ligolw.ilwd.sngl_ringdown_event_id_class object>¶
-
tableName= 'sngl_ringdown:table'¶
-
validcolumns= {'ds2_H1H2': 'real_4', 'num_clust_trigs': 'int_4s', 'frequency': 'real_4', 'ds2_L1V1': 'real_4', 'quality': 'real_4', 'event_id': 'ilwd:char', 'spin': 'real_4', 'sigma_sq': 'real_8', 'channel': 'lstring', 'ds2_H2L1': 'real_4', 'epsilon': 'real_4', 'start_time': 'int_4s', 'ds2_H2V1': 'real_4', 'snr': 'real_4', 'start_time_gmst': 'real_8', 'phase': 'real_4', 'ds2_H1V1': 'real_4', 'start_time_ns': 'int_4s', 'eff_dist': 'real_4', 'process_id': 'ilwd:char', 'mass': 'real_4', 'amplitude': 'real_4', 'ifo': 'lstring', 'ds2_H1L1': 'real_4'}¶
Methods Documentation
-
binned_event_rates(stride, column, bins, operator='>=', start=None, end=None, timecolumn='time')[source]¶ Calculate an event rate
TimeSeriesDictover a number of bins.Parameters: stride :
floatsize (seconds) of each time bin
column :
strname of column by which to bin.
bins :
listone of:
'<','<=','>','>=','==','!=', for a standard mathematical operation,'in'to use the list of bins as containing bin edges, or- a callable function that takes compares an event value against the bin value and returns a boolean.
Note
If
binsis given as a list of tuples, this argument is ignored.start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeries.end :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rates :
TimeSeriesDicta dict of (bin,
TimeSeries) pairs describing a rate of events per second (Hz) for each of the bins.
-
event_rate(stride, start=None, end=None, timecolumn='time')[source]¶ Calculate the rate
TimeSeriesfor thisTable.Parameters: stride :
floatsize (seconds) of each time bin
start :
float,LIGOTimeGPS, optionalGPS start epoch of rate
TimeSeriesend :
float,LIGOTimeGPS, optionalGPS end time of rate
TimeSeries. This value will be rounded up to the nearest sample if needed.timecolumn :
str, optional, default:timename of time-column to use when binning events
Returns: rate :
TimeSeriesa
TimeSeriesof events per second (Hz)
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_start()¶
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SnglRingdownTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SnglRingdownTableSnglRingdownTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes sngl_ringdown Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.StochasticTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
tableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a StochasticTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
tableName= 'stochastic:table'¶
-
validcolumns= {'channel_two': 'lstring', 'start_time': 'int_4s', 'channel_one': 'lstring', 'duration_ns': 'int_4s', 'process_id': 'ilwd:char', 'cc_stat': 'real_8', 'duration': 'int_4s', 'f_max': 'real_8', 'ifo_two': 'lstring', 'start_time_ns': 'int_4s', 'cc_sigma': 'real_8', 'f_min': 'real_8', 'ifo_one': 'lstring'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
StochasticTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
StochasticTableStochasticTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes stochastic Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.StochSummTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
tableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a StochSummTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
tableName= 'stochsumm:table'¶
-
validcolumns= {'channel_two': 'lstring', 'start_time': 'int_4s', 'channel_one': 'lstring', 'process_id': 'ilwd:char', 'f_max': 'real_8', 'ifo_two': 'lstring', 'start_time_ns': 'int_4s', 'f_min': 'real_8', 'ifo_one': 'lstring', 'end_time': 'int_4s', 'error': 'real_8', 'end_time_ns': 'int_4s', 'y_opt': 'real_8'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
StochSummTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
StochSummTableStochSummTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes stochsumm Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SummValueTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a SummValueTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (summ_value_id)'¶
-
interncolumns= ('program', 'process_id', 'ifo', 'name', 'comment')¶
-
next_id= <glue.ligolw.ilwd.summ_value_summ_value_id_class object>¶
-
tableName= 'summ_value:table'¶
-
validcolumns= {'comment': 'lstring', 'start_time': 'int_4s', 'process_id': 'ilwd:char', 'summ_value_id': 'ilwd:char', 'segment_def_id': 'ilwd:char', 'ifo': 'lstring', 'name': 'lstring', 'start_time_ns': 'int_4s', 'value': 'real_4', 'intvalue': 'int_4s', 'program': 'lstring', 'end_time': 'int_4s', 'error': 'real_4', 'end_time_ns': 'int_4s', 'frameset_group': 'lstring'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SummValueTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SummValueTableSummValueTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes summ_value Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SegmentTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a SegmentTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (segment_id)'¶
-
interncolumns= ('process_id',)¶
-
next_id= <glue.ligolw.ilwd.segment_segment_id_class object>¶
-
tableName= 'segment:table'¶
-
validcolumns= {'segment_def_cdb': 'int_4s', 'process_id': 'ilwd:char', 'creator_db': 'int_4s', 'end_time': 'int_4s', 'segment_id': 'ilwd:char', 'segment_def_id': 'ilwd:char', 'start_time': 'int_4s', 'start_time_ns': 'int_4s', 'end_time_ns': 'int_4s'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SegmentTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SegmentTableSegmentTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes segment Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SegmentSumTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayget([segment_def_id])Return a segmentlist object describing the times spanned by the segments carrying the given segment_def_id. hist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a SegmentSumTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (segment_sum_id)'¶
-
interncolumns= ('process_id', 'segment_def_id')¶
-
next_id= <glue.ligolw.ilwd.segment_summary_segment_sum_id_class object>¶
-
tableName= 'segment_summary:table'¶
-
validcolumns= {'comment': 'lstring', 'segment_def_cdb': 'int_4s', 'process_id': 'ilwd:char', 'creator_db': 'int_4s', 'end_time': 'int_4s', 'start_time_ns': 'int_4s', 'segment_def_id': 'ilwd:char', 'start_time': 'int_4s', 'end_time_ns': 'int_4s', 'segment_sum_id': 'ilwd:char'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get(segment_def_id=None)¶ Return a segmentlist object describing the times spanned by the segments carrying the given segment_def_id. If segment_def_id is None then all segments are returned.
Note: the result is not coalesced, the segmentlist contains the segments as they appear in the table.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SegmentSumTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SegmentSumTableSegmentSumTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes segment_summary Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SegmentDefTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayread(*args, **kwargs)Read data into a SegmentDefTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (segment_def_id)'¶
-
interncolumns= ('process_id',)¶
-
next_id= <glue.ligolw.ilwd.segment_definer_segment_def_id_class object>¶
-
tableName= 'segment_definer:table'¶
-
validcolumns= {'comment': 'lstring', 'process_id': 'ilwd:char', 'creator_db': 'int_4s', 'name': 'lstring', 'version': 'int_4s', 'segment_def_id': 'ilwd:char', 'insertion_time': 'int_4s', 'ifos': 'lstring'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SegmentDefTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SegmentDefTableSegmentDefTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes segment_definer Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.SummMimeTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsnext_idtableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a SummMimeTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (summ_mime_id)'¶
-
next_id= <glue.ligolw.ilwd.summ_mime_summ_mime_id_class object>¶
-
tableName= 'summ_mime:table'¶
-
validcolumns= {'origin': 'lstring', 'mimedata': 'blob', 'start_time_ns': 'int_4s', 'start_time': 'int_4s', 'summ_mime_id': 'ilwd:char', 'comment': 'lstring', 'filename': 'lstring', 'mimedata_length': 'int_4s', 'process_id': 'ilwd:char', 'end_time': 'int_4s', 'mimetype': 'lstring', 'descrip': 'lstring', 'submitter': 'lstring', 'segment_def_id': 'ilwd:char', 'frameset_group': 'lstring', 'end_time_ns': 'int_4s', 'channel': 'lstring'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
SummMimeTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
SummMimeTableSummMimeTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes summ_mime Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.TimeSlideSegmentMapTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
tableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayread(*args, **kwargs)Read data into a TimeSlideSegmentMapTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
tableName= 'time_slide_segment_map:table'¶
-
validcolumns= {'time_slide_id': 'ilwd:char', 'segment_def_id': 'ilwd:char'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
classmethod
read(*args, **kwargs)¶ Read data into a
TimeSlideSegmentMapTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
TimeSlideSegmentMapTableTimeSlideSegmentMapTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes time_slide_segment_map Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.TimeSlideTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
constraintsinterncolumnsnext_idtableNamevalidcolumnsMethods Summary
append_offsetvector(offsetvect, process)Append rows describing an instrument –> offset mapping to this table. as_dict()Return a ditionary mapping time slide IDs to offset dictionaries. from_recarray(array[, columns])Create a new table from a numpy.recarrayget_time_slide_id(offsetdict[, create_new, ...])Return the time_slide_id corresponding to the offset vector described by offsetdict, a dictionary of instrument/offset pairs. read(*args, **kwargs)Read data into a TimeSlideTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
constraints= 'PRIMARY KEY (time_slide_id, instrument)'¶
-
interncolumns= ('process_id', 'time_slide_id', 'instrument')¶
-
next_id= <glue.ligolw.ilwd.time_slide_time_slide_id_class object>¶
-
tableName= 'time_slide:table'¶
-
validcolumns= {'instrument': 'lstring', 'time_slide_id': 'ilwd:char', 'process_id': 'ilwd:char', 'offset': 'real_8'}¶
Methods Documentation
-
append_offsetvector(offsetvect, process)¶ Append rows describing an instrument –> offset mapping to this table. offsetvect is a dictionary mapping instrument to offset. process should be the row in the process table on which the new time_slide table rows will be blamed (or any object with a process_id attribute). The return value is the time_slide_id assigned to the new rows.
-
as_dict()¶ Return a ditionary mapping time slide IDs to offset dictionaries.
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
get_time_slide_id(offsetdict, create_new=None, superset_ok=False, nonunique_ok=False)¶ Return the time_slide_id corresponding to the offset vector described by offsetdict, a dictionary of instrument/offset pairs.
If the optional create_new argument is None (the default), then the table must contain a matching offset vector. The return value is the ID of that vector. If the table does not contain a matching offset vector then KeyError is raised.
If the optional create_new argument is set to a Process object (or any other object with a process_id attribute), then if the table does not contain a matching offset vector a new one will be added to the table and marked as having been created by the given process. The return value is the ID of the (possibly newly created) matching offset vector.
If the optional superset_ok argument is False (the default) then an offset vector in the table is considered to “match” the requested offset vector only if they contain the exact same set of instruments. If the superset_ok argument is True, then an offset vector in the table is considered to match the requested offset vector as long as it provides the same offsets for the same instruments as the requested vector, even if it provides offsets for other instruments as well.
More than one offset vector in the table might match the requested vector. If the optional nonunique_ok argument is False (the default), then KeyError will be raised if more than one offset vector in the table is found to match the requested vector. If the optional nonunique_ok is True then the return value is the ID of one of the matching offset vectors selected at random.
-
classmethod
read(*args, **kwargs)¶ Read data into a
TimeSlideTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
TimeSlideTableTimeSlideTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes time_slide Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-
-
class
gwpy.table.lsctables.VetoDefTable(*args)¶ Bases:
glue.ligolw.table.TableAttributes Summary
interncolumnstableNamevalidcolumnsMethods Summary
from_recarray(array[, columns])Create a new table from a numpy.recarrayhist(column, **kwargs)Generate a HistogramPlotof thisTable.plot(*args, **kwargs)Generate an EventTablePlotof thisTable.read(*args, **kwargs)Read data into a VetoDefTable.to_recarray([columns, on_attributeerror])Convert this table to a structured numpy.recarrayAttributes Documentation
-
interncolumns= ('process_id', 'ifo')¶
-
tableName= 'veto_definer:table'¶
-
validcolumns= {'category': 'int_4s', 'comment': 'lstring', 'end_pad': 'int_4s', 'process_id': 'ilwd:char', 'name': 'lstring', 'version': 'int_4s', 'start_pad': 'int_4s', 'start_time': 'int_4s', 'ifo': 'lstring', 'end_time': 'int_4s'}¶
Methods Documentation
-
classmethod
from_recarray(array, columns=None)[source]¶ Create a new table from a
numpy.recarrayParameters: array :
numpy.recarrayan array of data
column :
listofstr, optionalthe columns to populate, if not given, all columns present in the
recarrayare mappedNotes
The columns populated in the
numpy.recarraymust all map exactly to valid columns of the targetTable.
-
hist(column, **kwargs)[source]¶ Generate a
HistogramPlotof thisTable.Parameters: column :
strname of the column over which to histogram data
**kwargs :
any other arguments applicable to the
HistogramPlotReturns: plot :
HistogramPlotnew plot displaying a histogram of this
Table.
-
plot(*args, **kwargs)[source]¶ Generate an
EventTablePlotof thisTable.Parameters: x :
strname of column defining centre point on the X-axis
y :
strname of column defining centre point on the Y-axis
width :
str, optionalname of column defining width of tile
height :
str, optionalname of column defining height of tile
Note
The
widthandheightpositional arguments should either both not given, in which case a scatter plot will be drawn, or both given, in which case a collections of rectangles will be drawn.color :
str, optional, default:Nonename of column by which to color markers
**kwargs :
any other arguments applicable to the
Plotconstructor, and theTableplotter.Returns: plot :
EventTablePlotnew plot for displaying tabular data.
See also
gwpy.plotter.EventTablePlot- for more details.
-
classmethod
read(*args, **kwargs)¶ Read data into a
VetoDefTable.Parameters: f :
file,str,CacheEntry,list,Cachecolumns :
list, optionallist of column name strings to read, default all.
ifo :
str, optionalprefix of IFO to read
Warning
the
ifokeyword argument is only applicable (but is required) when reading single-interferometer data from a multi-interferometer filefilt :
function, optionalnproc :
int, optional, default:1contenthandler :
LIGOLWContentHandlerSAX content handler for parsing
LIGO_LWdocuments.Warning
The
contenthandlerkeyword argument is only applicable when reading fromLIGO_LWdocuments.**loadtxtkwargs :
when reading from ASCII, all other keyword arguments are passed directly to
numpy.loadtxtReturns: table :
VetoDefTableVetoDefTableof data with given columns filledNotes
The available built-in formats are:
Format Read Write Auto-identify ascii Yes No Yes csv Yes No No ligolw Yes No Yes veto_definer Yes No No
-
to_recarray(columns=None, on_attributeerror='raise')[source]¶ Convert this table to a structured
numpy.recarrayThis returned
recarrayis a blank data container, mapping columns in the original LIGO_LW table to fields in the output, but mapping none of the instance methods of the origin table.Parameters: columns :
listofstr, optionalthe columns to populate, if not given, all columns present in the table are mapped
on_attributeerror :
strhow to handle
AttributeErrorwhen accessing rows, one of- ‘raise’ : raise normal exception
- ‘ignore’ : skip over this column
- ‘warn’ : print a warning instead of raising error
-