Spectrogram¶
-
class
gwpy.spectrogram.Spectrogram[source]¶ Bases:
gwpy.types.array2d.Array2DA 2D array holding a spectrogram of time-frequency data
- Parameters
value : array-like
input data array
unit :
Unit, optionalphysical unit of these data
epoch :
LIGOTimeGPS,float,str, optionalGPS epoch associated with these data, any input parsable by
to_gpsis finesample_rate :
float,Quantity, optional, default:1the rate of samples per second (Hertz)
times :
array-likethe complete array of GPS times accompanying the data for this series. This argument takes precedence over
epochandsample_rateso should be given in place of these if relevant, not alongsidef0 :
float,Quantity, optional, default:0starting frequency for these data
df :
float,Quantity, optional, default:1frequency resolution for these data
frequencies :
array-likeepoch :
LIGOTimeGPS,float,str, optionalGPS epoch associated with these data, any input parsable by
to_gpsis finename :
str, optionaldescriptive title for this array
channel :
Channel,str, optionalsource data stream for these data
dtype :
dtype, optionalinput data type
copy :
bool, optional, default:Falsechoose to copy the input data to new memory
subok :
bool, optional, default:Trueallow passing of sub-classes by the array generator
Notes
Key methods:
read(source, \*args, \*\*kwargs)Read data into a
Spectrogramwrite(self, target, \*args, \*\*kwargs)Write this
Spectrogramto a fileplot(self[, figsize, xscale])Plot the data for this
Spectrogramzpk(self, zeros, poles, gain[, analog])Filter this
Spectrogramby applying a zero-pole-gain filterAttributes Summary
The transposed array.
Frequency band described by this
SpectrogramBase object if memory is from some other object.
Returns a copy of the current
Quantityinstance with CGS units.Instrumental channel associated with these data
An object to simplify the interaction of the array with the ctypes module.
Python buffer object pointing to the start of the array’s data.
Frequency spacing of this
SpectrogramTime-spacing for this
SpectrogramData-type of the array’s elements.
X-axis sample separation
Y-axis sample separation
Starting GPS epoch for this
SpectrogramA list of equivalencies that will be applied by default during unit conversions.
Starting frequency for this
SpectrogramInformation about the memory layout of the array.
A 1-D iterator over the Quantity array.
Series of frequencies for this Spectrogram
The imaginary part of the array.
info([option, out])Container for meta information like name, description, format.
True if the
valueof this quantity is a scalar, or False if it is an array-like object.Length of one array element in bytes.
Name for this data set
Total bytes consumed by the elements of the array.
Number of array dimensions.
The real part of the array.
Tuple of array dimensions.
Returns a copy of the current
Quantityinstance with SI units.Number of elements in the array.
GPS [start, stop) span for this
SpectrogramTuple of bytes to step in each dimension when traversing an array.
GPS time of first time bin
Series of GPS times for each sample
The physical unit of these data
The numerical value of this instance.
X-axis coordinate of the first data point
Positions of the data on the x-axis
X-axis [low, high) segment encompassed by these data
Unit of x-axis index
Y-axis coordinate of the first data point
Positions of the data on the y-axis
Y-axis [low, high) segment encompassed by these data
Unit of Y-axis index
Methods Summary
abs(x, /[, out, where, casting, order, …])Calculate the absolute value element-wise.
all([axis, out, keepdims])Returns True if all elements evaluate to True.
any([axis, out, keepdims])Returns True if any of the elements of
aevaluate to True.append(self, other[, inplace, pad, gap, resize])Connect another series onto the end of the current one.
argmax([axis, out])Return indices of the maximum values along the given axis.
argmin([axis, out])Return indices of the minimum values along the given axis of
a.argpartition(kth[, axis, kind, order])Returns the indices that would partition this array.
argsort([axis, kind, order])Returns the indices that would sort this array.
astype(dtype[, order, casting, subok, copy])Copy of the array, cast to a specified type.
byteswap([inplace])Swap the bytes of the array elements
choose(choices[, out, mode])Use an index array to construct a new array from a set of choices.
clip([min, max, out])Return an array whose values are limited to
[min, max].compress(condition[, axis, out])Return selected slices of this array along given axis.
conj()Complex-conjugate all elements.
Return the complex conjugate, element-wise.
copy([order])Return a copy of the array.
crop(self[, start, end, copy])Crop this series to the given x-axis extent.
crop_frequencies(self[, low, high, copy])Crop this
Spectrogramto the specified frequenciescumprod([axis, dtype, out])Return the cumulative product of the elements along the given axis.
cumsum([axis, dtype, out])Return the cumulative sum of the elements along the given axis.
decompose(self[, bases])Generates a new
Quantitywith the units decomposed.diagonal([offset, axis1, axis2])Return specified diagonals.
diff(self[, n, axis])Calculate the n-th order discrete difference along given axis.
dot(b[, out])Dot product of two arrays.
dump(file)Dump a pickle of the array to the specified file.
dumps()Returns the pickle of the array as a string.
ediff1d(self[, to_end, to_begin])fill(value)Fill the array with a scalar value.
filter(self, \*filt, \*\*kwargs)Apply the given filter to this
Spectrogram.flatten(self[, order])Return a copy of the array collapsed into one dimension.
from_spectra(\*spectra, \*\*kwargs)Build a new
Spectrogramfrom a list of spectra.getfield(dtype[, offset])Returns a field of the given array as a certain type.
imshow(self, \*\*kwargs)inject(self, other)Add two compatible
Seriesalong their shared x-axis values.insert(self, obj, values[, axis])Insert values along the given axis before the given indices and return a new
Quantityobject.is_compatible(self, other)Check whether this array and
otherhave compatible metadatais_contiguous(self, other[, tol])Check whether other is contiguous with self.
item(*args)Copy an element of an array to a standard Python scalar and return it.
itemset(*args)Insert scalar into an array (scalar is cast to array’s dtype, if possible)
max([axis, out, keepdims, initial, where])Return the maximum along a given axis.
mean([axis, dtype, out, keepdims])Returns the average of the array elements along given axis.
median(self[, axis])Compute the median along the specified axis.
min([axis, out, keepdims, initial, where])Return the minimum along a given axis.
nansum(self[, axis, out, keepdims])newbyteorder([new_order])Return the array with the same data viewed with a different byte order.
nonzero()Return the indices of the elements that are non-zero.
override_unit(self, unit[, parse_strict])Forcefully reset the unit of these data
pad(self, pad_width, \*\*kwargs)Pad this series to a new size
partition(kth[, axis, kind, order])Rearranges the elements in the array in such a way that the value of the element in kth position is in the position it would be in a sorted array.
pcolormesh(self, \*\*kwargs)percentile(self, percentile)Calculate a given spectral percentile for this
Spectrogram.plot(self[, figsize, xscale])Plot the data for this
Spectrogramprepend(self, other[, inplace, pad, gap, resize])Connect another series onto the start of the current one.
prod([axis, dtype, out, keepdims, initial, …])Return the product of the array elements over the given axis
ptp([axis, out, keepdims])Peak to peak (maximum - minimum) value along a given axis.
put(indices, values[, mode])Set
a.flat[n] = values[n]for allnin indices.ratio(self, operand)Calculate the ratio of this
Spectrogramagainst a referenceravel([order])Return a flattened array.
read(source, \*args, \*\*kwargs)Read data into a
Spectrogramrepeat(repeats[, axis])Repeat elements of an array.
reshape(shape[, order])Returns an array containing the same data with a new shape.
resize(new_shape[, refcheck])Change shape and size of array in-place.
round([decimals, out])Return
awith each element rounded to the given number of decimals.searchsorted(v[, side, sorter])Find indices where elements of v should be inserted in a to maintain order.
setfield(val, dtype[, offset])Put a value into a specified place in a field defined by a data-type.
setflags([write, align, uic])Set array flags WRITEABLE, ALIGNED, (WRITEBACKIFCOPY and UPDATEIFCOPY), respectively.
shift(self, delta)Shift this
Seriesforward on the X-axis bydeltasort([axis, kind, order])Sort an array in-place.
squeeze([axis])Remove single-dimensional entries from the shape of
a.std([axis, dtype, out, ddof, keepdims])Returns the standard deviation of the array elements along given axis.
step(self, \*\*kwargs)Create a step plot of this series
sum([axis, dtype, out, keepdims, initial, where])Return the sum of the array elements over the given axis.
swapaxes(axis1, axis2)Return a view of the array with
axis1andaxis2interchanged.take(indices[, axis, out, mode])Return an array formed from the elements of
aat the given indices.to(self, unit[, equivalencies])Return a new
Quantityobject with the specified unit.to_string(self[, unit, precision, format, …])Generate a string representation of the quantity and its unit.
to_value(self[, unit, equivalencies])The numerical value, possibly in a different unit.
tobytes([order])Construct Python bytes containing the raw data bytes in the array.
tofile(fid[, sep, format])Write array to a file as text or binary (default).
tolist()Return the array as an
a.ndim-levels deep nested list of Python scalars.tostring([order])Construct Python bytes containing the raw data bytes in the array.
trace([offset, axis1, axis2, dtype, out])Return the sum along diagonals of the array.
transpose(*axes)Returns a view of the array with axes transposed.
update(self, other[, inplace])Update this series by appending new data from an other and dropping the same amount of data off the start.
value_at(self, x, y)Return the value of this
Seriesat the given(x, y)coordinatesvar([axis, dtype, out, ddof, keepdims])Returns the variance of the array elements, along given axis.
variance(self[, bins, low, high, nbins, …])Calculate the
SpectralVarianceof thisSpectrogram.view([dtype, type])New view of array with the same data.
write(self, target, \*args, \*\*kwargs)Write this
Spectrogramto a filezip(self)zpk(self, zeros, poles, gain[, analog])Filter this
Spectrogramby applying a zero-pole-gain filterAttributes Documentation
-
T¶ The transposed array.
Same as
self.transpose().See also
Examples
>>> x = np.array([[1.,2.],[3.,4.]]) >>> x array([[ 1., 2.], [ 3., 4.]]) >>> x.T array([[ 1., 3.], [ 2., 4.]]) >>> x = np.array([1.,2.,3.,4.]) >>> x array([ 1., 2., 3., 4.]) >>> x.T array([ 1., 2., 3., 4.])
-
band¶ Frequency band described by this
Spectrogram
-
base¶ Base object if memory is from some other object.
Examples
The base of an array that owns its memory is None:
>>> x = np.array([1,2,3,4]) >>> x.base is None True
Slicing creates a view, whose memory is shared with x:
>>> y = x[2:] >>> y.base is x True
-
cgs¶ Returns a copy of the current
Quantityinstance with CGS units. The value of the resulting object will be scaled.
-
ctypes¶ An object to simplify the interaction of the array with the ctypes module.
This attribute creates an object that makes it easier to use arrays when calling shared libraries with the ctypes module. The returned object has, among others, data, shape, and strides attributes (see Notes below) which themselves return ctypes objects that can be used as arguments to a shared library.
- Parameters
- None
- Returns
c : Python object
Possessing attributes data, shape, strides, etc.
See also
Notes
Below are the public attributes of this object which were documented in “Guide to NumPy” (we have omitted undocumented public attributes, as well as documented private attributes):
-
_ctypes.data A pointer to the memory area of the array as a Python integer. This memory area may contain data that is not aligned, or not in correct byte-order. The memory area may not even be writeable. The array flags and data-type of this array should be respected when passing this attribute to arbitrary C-code to avoid trouble that can include Python crashing. User Beware! The value of this attribute is exactly the same as
self._array_interface_['data'][0].Note that unlike
data_as, a reference will not be kept to the array: code likectypes.c_void_p((a + b).ctypes.data)will result in a pointer to a deallocated array, and should be spelt(a + b).ctypes.data_as(ctypes.c_void_p)
-
_ctypes.shape (c_intp*self.ndim): A ctypes array of length self.ndim where the basetype is the C-integer corresponding to
dtype('p')on this platform. This base-type could bectypes.c_int,ctypes.c_long, orctypes.c_longlongdepending on the platform. The c_intp type is defined accordingly innumpy.ctypeslib. The ctypes array contains the shape of the underlying array.
-
_ctypes.strides (c_intp*self.ndim): A ctypes array of length self.ndim where the basetype is the same as for the shape attribute. This ctypes array contains the strides information from the underlying array. This strides information is important for showing how many bytes must be jumped to get to the next element in the array.
-
_ctypes.data_as(self, obj) Return the data pointer cast to a particular c-types object. For example, calling
self._as_parameter_is equivalent toself.data_as(ctypes.c_void_p). Perhaps you want to use the data as a pointer to a ctypes array of floating-point data:self.data_as(ctypes.POINTER(ctypes.c_double)).The returned pointer will keep a reference to the array.
-
_ctypes.shape_as(self, obj) Return the shape tuple as an array of some other c-types type. For example:
self.shape_as(ctypes.c_short).
-
_ctypes.strides_as(self, obj) Return the strides tuple as an array of some other c-types type. For example:
self.strides_as(ctypes.c_longlong).
If the ctypes module is not available, then the ctypes attribute of array objects still returns something useful, but ctypes objects are not returned and errors may be raised instead. In particular, the object will still have the
as_parameterattribute which will return an integer equal to the data attribute.Examples
>>> import ctypes >>> x array([[0, 1], [2, 3]]) >>> x.ctypes.data 30439712 >>> x.ctypes.data_as(ctypes.POINTER(ctypes.c_long)) <ctypes.LP_c_long object at 0x01F01300> >>> x.ctypes.data_as(ctypes.POINTER(ctypes.c_long)).contents c_long(0) >>> x.ctypes.data_as(ctypes.POINTER(ctypes.c_longlong)).contents c_longlong(4294967296L) >>> x.ctypes.shape <numpy.core._internal.c_long_Array_2 object at 0x01FFD580> >>> x.ctypes.shape_as(ctypes.c_long) <numpy.core._internal.c_long_Array_2 object at 0x01FCE620> >>> x.ctypes.strides <numpy.core._internal.c_long_Array_2 object at 0x01FCE620> >>> x.ctypes.strides_as(ctypes.c_longlong) <numpy.core._internal.c_longlong_Array_2 object at 0x01F01300>
-
data¶ Python buffer object pointing to the start of the array’s data.
-
df¶ Frequency spacing of this
Spectrogram- Type
Quantityin Hertz
-
dt¶ Time-spacing for this
Spectrogram- Type
Quantityin seconds
-
dtype¶ Data-type of the array’s elements.
- Parameters
- None
- Returns
- dnumpy dtype object
See also
Examples
>>> x array([[0, 1], [2, 3]]) >>> x.dtype dtype('int32') >>> type(x.dtype) <type 'numpy.dtype'>
-
epoch¶ Starting GPS epoch for this
Spectrogram- Type
-
equivalencies¶ A list of equivalencies that will be applied by default during unit conversions.
-
f0¶ Starting frequency for this
Spectrogram- Type
Quantityin Hertz
-
flags¶ Information about the memory layout of the array.
Notes
The
flagsobject can be accessed dictionary-like (as ina.flags['WRITEABLE']), or by using lowercased attribute names (as ina.flags.writeable). Short flag names are only supported in dictionary access.Only the WRITEBACKIFCOPY, UPDATEIFCOPY, WRITEABLE, and ALIGNED flags can be changed by the user, via direct assignment to the attribute or dictionary entry, or by calling
ndarray.setflags.The array flags cannot be set arbitrarily:
UPDATEIFCOPY can only be set
False.WRITEBACKIFCOPY can only be set
False.ALIGNED can only be set
Trueif the data is truly aligned.WRITEABLE can only be set
Trueif the array owns its own memory or the ultimate owner of the memory exposes a writeable buffer interface or is a string.
Arrays can be both C-style and Fortran-style contiguous simultaneously. This is clear for 1-dimensional arrays, but can also be true for higher dimensional arrays.
Even for contiguous arrays a stride for a given dimension
arr.strides[dim]may be arbitrary ifarr.shape[dim] == 1or the array has no elements. It does not generally hold thatself.strides[-1] == self.itemsizefor C-style contiguous arrays orself.strides[0] == self.itemsizefor Fortran-style contiguous arrays is true.- Attributes
C_CONTIGUOUS (C)
The data is in a single, C-style contiguous segment.
F_CONTIGUOUS (F)
The data is in a single, Fortran-style contiguous segment.
OWNDATA (O)
The array owns the memory it uses or borrows it from another object.
WRITEABLE (W)
The data area can be written to. Setting this to False locks the data, making it read-only. A view (slice, etc.) inherits WRITEABLE from its base array at creation time, but a view of a writeable array may be subsequently locked while the base array remains writeable. (The opposite is not true, in that a view of a locked array may not be made writeable. However, currently, locking a base object does not lock any views that already reference it, so under that circumstance it is possible to alter the contents of a locked array via a previously created writeable view onto it.) Attempting to change a non-writeable array raises a RuntimeError exception.
ALIGNED (A)
The data and all elements are aligned appropriately for the hardware.
WRITEBACKIFCOPY (X)
This array is a copy of some other array. The C-API function PyArray_ResolveWritebackIfCopy must be called before deallocating to the base array will be updated with the contents of this array.
UPDATEIFCOPY (U)
(Deprecated, use WRITEBACKIFCOPY) This array is a copy of some other array. When this array is deallocated, the base array will be updated with the contents of this array.
FNC
F_CONTIGUOUS and not C_CONTIGUOUS.
FORC
F_CONTIGUOUS or C_CONTIGUOUS (one-segment test).
BEHAVED (B)
ALIGNED and WRITEABLE.
CARRAY (CA)
BEHAVED and C_CONTIGUOUS.
FARRAY (FA)
BEHAVED and F_CONTIGUOUS and not C_CONTIGUOUS.
-
flat¶ A 1-D iterator over the Quantity array.
This returns a
QuantityIteratorinstance, which behaves the same as theflatiterinstance returned byflat, and is similar to, but not a subclass of, Python’s built-in iterator object.
-
frequencies¶ Series of frequencies for this Spectrogram
-
imag¶ The imaginary part of the array.
Examples
>>> x = np.sqrt([1+0j, 0+1j]) >>> x.imag array([ 0. , 0.70710678]) >>> x.imag.dtype dtype('float64')
-
info(option='attributes', out='')¶ Container for meta information like name, description, format. This is required when the object is used as a mixin column within a table, but can be used as a general way to store meta information.
-
isscalar¶ True if the
valueof this quantity is a scalar, or False if it is an array-like object.Note
This is subtly different from
numpy.isscalarin thatnumpy.isscalarreturns False for a zero-dimensional array (e.g.np.array(1)), while this is True for quantities, since quantities cannot represent true numpy scalars.
-
itemsize¶ Length of one array element in bytes.
Examples
>>> x = np.array([1,2,3], dtype=np.float64) >>> x.itemsize 8 >>> x = np.array([1,2,3], dtype=np.complex128) >>> x.itemsize 16
-
nbytes¶ Total bytes consumed by the elements of the array.
Notes
Does not include memory consumed by non-element attributes of the array object.
Examples
>>> x = np.zeros((3,5,2), dtype=np.complex128) >>> x.nbytes 480 >>> np.prod(x.shape) * x.itemsize 480
-
ndim¶ Number of array dimensions.
Examples
>>> x = np.array([1, 2, 3]) >>> x.ndim 1 >>> y = np.zeros((2, 3, 4)) >>> y.ndim 3
-
real¶ The real part of the array.
See also
numpy.realequivalent function
Examples
>>> x = np.sqrt([1+0j, 0+1j]) >>> x.real array([ 1. , 0.70710678]) >>> x.real.dtype dtype('float64')
-
shape¶ Tuple of array dimensions.
The shape property is usually used to get the current shape of an array, but may also be used to reshape the array in-place by assigning a tuple of array dimensions to it. As with
numpy.reshape, one of the new shape dimensions can be -1, in which case its value is inferred from the size of the array and the remaining dimensions. Reshaping an array in-place will fail if a copy is required.See also
numpy.reshapesimilar function
ndarray.reshapesimilar method
Examples
>>> x = np.array([1, 2, 3, 4]) >>> x.shape (4,) >>> y = np.zeros((2, 3, 4)) >>> y.shape (2, 3, 4) >>> y.shape = (3, 8) >>> y array([[ 0., 0., 0., 0., 0., 0., 0., 0.], [ 0., 0., 0., 0., 0., 0., 0., 0.], [ 0., 0., 0., 0., 0., 0., 0., 0.]]) >>> y.shape = (3, 6) Traceback (most recent call last): File "<stdin>", line 1, in <module> ValueError: total size of new array must be unchanged >>> np.zeros((4,2))[::2].shape = (-1,) Traceback (most recent call last): File "<stdin>", line 1, in <module> AttributeError: incompatible shape for a non-contiguous array
-
si¶ Returns a copy of the current
Quantityinstance with SI units. The value of the resulting object will be scaled.
-
size¶ Number of elements in the array.
Equal to
np.prod(a.shape), i.e., the product of the array’s dimensions.Notes
a.sizereturns a standard arbitrary precision Python integer. This may not be the case with other methods of obtaining the same value (like the suggestednp.prod(a.shape), which returns an instance ofnp.int_), and may be relevant if the value is used further in calculations that may overflow a fixed size integer type.Examples
>>> x = np.zeros((3, 5, 2), dtype=np.complex128) >>> x.size 30 >>> np.prod(x.shape) 30
-
span¶ GPS [start, stop) span for this
Spectrogram- Type
-
strides¶ Tuple of bytes to step in each dimension when traversing an array.
The byte offset of element
(i[0], i[1], ..., i[n])in an arrayais:offset = sum(np.array(i) * a.strides)
A more detailed explanation of strides can be found in the “ndarray.rst” file in the NumPy reference guide.
See also
Notes
Imagine an array of 32-bit integers (each 4 bytes):
x = np.array([[0, 1, 2, 3, 4], [5, 6, 7, 8, 9]], dtype=np.int32)
This array is stored in memory as 40 bytes, one after the other (known as a contiguous block of memory). The strides of an array tell us how many bytes we have to skip in memory to move to the next position along a certain axis. For example, we have to skip 4 bytes (1 value) to move to the next column, but 20 bytes (5 values) to get to the same position in the next row. As such, the strides for the array
xwill be(20, 4).Examples
>>> y = np.reshape(np.arange(2*3*4), (2,3,4)) >>> y array([[[ 0, 1, 2, 3], [ 4, 5, 6, 7], [ 8, 9, 10, 11]], [[12, 13, 14, 15], [16, 17, 18, 19], [20, 21, 22, 23]]]) >>> y.strides (48, 16, 4) >>> y[1,1,1] 17 >>> offset=sum(y.strides * np.array((1,1,1))) >>> offset/y.itemsize 17
>>> x = np.reshape(np.arange(5*6*7*8), (5,6,7,8)).transpose(2,3,1,0) >>> x.strides (32, 4, 224, 1344) >>> i = np.array([3,5,2,2]) >>> offset = sum(i * x.strides) >>> x[3,5,2,2] 813 >>> offset / x.itemsize 813
-
times¶ Series of GPS times for each sample
-
value¶ The numerical value of this instance.
See also
to_valueGet the numerical value in a given unit.
Methods Documentation
-
abs(x, /, out=None, *, where=True, casting='same_kind', order='K', dtype=None, subok=True[, signature, extobj])[source]¶ Calculate the absolute value element-wise.
np.absis a shorthand for this function.- Parameters
x : array_like
Input array.
out : ndarray, None, or tuple of ndarray and None, optional
A location into which the result is stored. If provided, it must have a shape that the inputs broadcast to. If not provided or
None, a freshly-allocated array is returned. A tuple (possible only as a keyword argument) must have length equal to the number of outputs.where : array_like, optional
This condition is broadcast over the input. At locations where the condition is True, the
outarray will be set to the ufunc result. Elsewhere, theoutarray will retain its original value. Note that if an uninitializedoutarray is created via the defaultout=None, locations within it where the condition is False will remain uninitialized.**kwargs
For other keyword-only arguments, see the ufunc docs.
- Returns
absolute : ndarray
An ndarray containing the absolute value of each element in
x. For complex input,a + ib, the absolute value is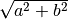 .
This is a scalar if
.
This is a scalar if xis a scalar.
Examples
>>> x = np.array([-1.2, 1.2]) >>> np.absolute(x) array([ 1.2, 1.2]) >>> np.absolute(1.2 + 1j) 1.5620499351813308
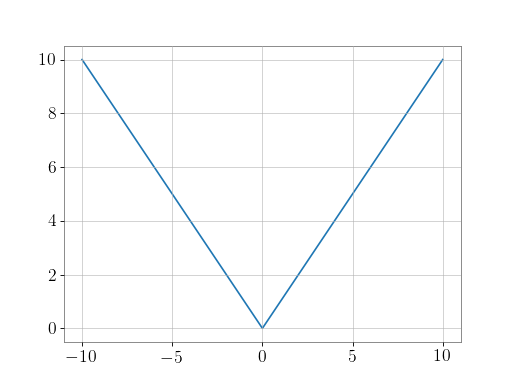
Plot the function over
[-10, 10]:>>> import matplotlib.pyplot as plt
>>> x = np.linspace(start=-10, stop=10, num=101) >>> plt.plot(x, np.absolute(x)) >>> plt.show()
(png)
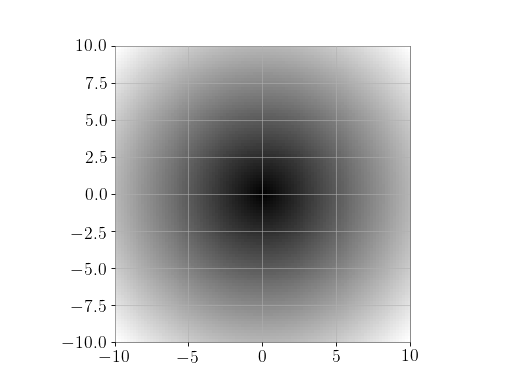
Plot the function over the complex plane:
>>> xx = x + 1j * x[:, np.newaxis] >>> plt.imshow(np.abs(xx), extent=[-10, 10, -10, 10], cmap='gray') >>> plt.show()
(png)
-
all(axis=None, out=None, keepdims=False)¶ Returns True if all elements evaluate to True.
Refer to
numpy.allfor full documentation.See also
numpy.allequivalent function
-
any(axis=None, out=None, keepdims=False)¶ Returns True if any of the elements of
aevaluate to True.Refer to
numpy.anyfor full documentation.See also
numpy.anyequivalent function
-
append(self, other, inplace=True, pad=None, gap=None, resize=True)[source]¶ Connect another series onto the end of the current one.
- Parameters
other :
Seriesanother series of the same type to connect to this one
inplace :
bool, optionalperform operation in-place, modifying current series, otherwise copy data and return new series, default:
TrueWarning
inplaceappend bypasses the reference check innumpy.ndarray.resize, so be carefully to only use this for arrays that haven’t been sharing their memory!pad :
float, optionalvalue with which to pad discontiguous series, by default gaps will result in a
ValueError.gap :
str, optionalaction to perform if there’s a gap between the other series and this one. One of
'raise'- raise aValueError'ignore'- remove gap and join data'pad'- pad gap with zeros
If
padis given and is notNone, the default is'pad', otherwise'raise'. Ifgap='pad'is given, the default forpadis0.resize :
bool, optionalresize this array to accommodate new data, otherwise shift the old data to the left (potentially falling off the start) and put the new data in at the end, default:
True.- Returns
series :
Seriesa new series containing joined data sets
-
argmax(axis=None, out=None)¶ Return indices of the maximum values along the given axis.
Refer to
numpy.argmaxfor full documentation.See also
numpy.argmaxequivalent function
-
argmin(axis=None, out=None)¶ Return indices of the minimum values along the given axis of
a.Refer to
numpy.argminfor detailed documentation.See also
numpy.argminequivalent function
-
argpartition(kth, axis=-1, kind='introselect', order=None)¶ Returns the indices that would partition this array.
Refer to
numpy.argpartitionfor full documentation.New in version 1.8.0.
See also
numpy.argpartitionequivalent function
-
argsort(axis=-1, kind=None, order=None)¶ Returns the indices that would sort this array.
Refer to
numpy.argsortfor full documentation.See also
numpy.argsortequivalent function
-
astype(dtype, order='K', casting='unsafe', subok=True, copy=True)¶ Copy of the array, cast to a specified type.
- Parameters
dtype : str or dtype
Typecode or data-type to which the array is cast.
order : {‘C’, ‘F’, ‘A’, ‘K’}, optional
Controls the memory layout order of the result. ‘C’ means C order, ‘F’ means Fortran order, ‘A’ means ‘F’ order if all the arrays are Fortran contiguous, ‘C’ order otherwise, and ‘K’ means as close to the order the array elements appear in memory as possible. Default is ‘K’.
casting : {‘no’, ‘equiv’, ‘safe’, ‘same_kind’, ‘unsafe’}, optional
Controls what kind of data casting may occur. Defaults to ‘unsafe’ for backwards compatibility.
‘no’ means the data types should not be cast at all.
‘equiv’ means only byte-order changes are allowed.
‘safe’ means only casts which can preserve values are allowed.
‘same_kind’ means only safe casts or casts within a kind, like float64 to float32, are allowed.
‘unsafe’ means any data conversions may be done.
subok : bool, optional
If True, then sub-classes will be passed-through (default), otherwise the returned array will be forced to be a base-class array.
copy : bool, optional
By default, astype always returns a newly allocated array. If this is set to false, and the
dtype,order, andsubokrequirements are satisfied, the input array is returned instead of a copy.- Returns
arr_t : ndarray
- Raises
ComplexWarning
When casting from complex to float or int. To avoid this, one should use
a.real.astype(t).
Notes
Changed in version 1.17.0: Casting between a simple data type and a structured one is possible only for “unsafe” casting. Casting to multiple fields is allowed, but casting from multiple fields is not.
Changed in version 1.9.0: Casting from numeric to string types in ‘safe’ casting mode requires that the string dtype length is long enough to store the max integer/float value converted.
Examples
>>> x = np.array([1, 2, 2.5]) >>> x array([1. , 2. , 2.5])
>>> x.astype(int) array([1, 2, 2])
-
byteswap(inplace=False)¶ Swap the bytes of the array elements
Toggle between low-endian and big-endian data representation by returning a byteswapped array, optionally swapped in-place.
- Parameters
inplace : bool, optional
If
True, swap bytes in-place, default isFalse.- Returns
out : ndarray
The byteswapped array. If
inplaceisTrue, this is a view to self.
Examples
>>> A = np.array([1, 256, 8755], dtype=np.int16) >>> list(map(hex, A)) ['0x1', '0x100', '0x2233'] >>> A.byteswap(inplace=True) array([ 256, 1, 13090], dtype=int16) >>> list(map(hex, A)) ['0x100', '0x1', '0x3322']
Arrays of strings are not swapped
>>> A = np.array(['ceg', 'fac']) >>> A.byteswap() Traceback (most recent call last): ... UnicodeDecodeError: ...
-
choose(choices, out=None, mode='raise')¶ Use an index array to construct a new array from a set of choices.
Refer to
numpy.choosefor full documentation.See also
numpy.chooseequivalent function
-
clip(min=None, max=None, out=None, **kwargs)¶ Return an array whose values are limited to
[min, max]. One of max or min must be given.Refer to
numpy.clipfor full documentation.See also
numpy.clipequivalent function
-
compress(condition, axis=None, out=None)¶ Return selected slices of this array along given axis.
Refer to
numpy.compressfor full documentation.See also
numpy.compressequivalent function
-
conj()¶ Complex-conjugate all elements.
Refer to
numpy.conjugatefor full documentation.See also
numpy.conjugateequivalent function
-
conjugate()¶ Return the complex conjugate, element-wise.
Refer to
numpy.conjugatefor full documentation.See also
numpy.conjugateequivalent function
-
copy(order='C')[source]¶ Return a copy of the array.
- Parameters
order : {‘C’, ‘F’, ‘A’, ‘K’}, optional
Controls the memory layout of the copy. ‘C’ means C-order, ‘F’ means F-order, ‘A’ means ‘F’ if
ais Fortran contiguous, ‘C’ otherwise. ‘K’ means match the layout ofaas closely as possible. (Note that this function andnumpy.copy()are very similar, but have different default values for their order= arguments.)
See also
Examples
>>> x = np.array([[1,2,3],[4,5,6]], order='F')
>>> y = x.copy()
>>> x.fill(0)
>>> x array([[0, 0, 0], [0, 0, 0]])
>>> y array([[1, 2, 3], [4, 5, 6]])
>>> y.flags['C_CONTIGUOUS'] True
-
crop(self, start=None, end=None, copy=False)[source]¶ Crop this series to the given x-axis extent.
- Parameters
start :
float, optionallower limit of x-axis to crop to, defaults to current
x0end :
float, optionalupper limit of x-axis to crop to, defaults to current series end
copy :
bool, optional, default:Falsecopy the input data to fresh memory, otherwise return a view
- Returns
series :
SeriesA new series with a sub-set of the input data
Notes
If either
startorendare outside of the originalSeriesspan, warnings will be printed and the limits will be restricted to thexspan
-
crop_frequencies(self, low=None, high=None, copy=False)[source]¶ Crop this
Spectrogramto the specified frequencies- Parameters
low :
floatlower frequency bound for cropped
Spectrogramhigh :
floatupper frequency bound for cropped
Spectrogramcopy :
boolif
Falsereturn a view of the original data, otherwise create a fresh memory copy- Returns
spec :
SpectrogramA new
Spectrogramwith a subset of data from the frequency axis
-
cumprod(axis=None, dtype=None, out=None)¶ Return the cumulative product of the elements along the given axis.
Refer to
numpy.cumprodfor full documentation.See also
numpy.cumprodequivalent function
-
cumsum(axis=None, dtype=None, out=None)¶ Return the cumulative sum of the elements along the given axis.
Refer to
numpy.cumsumfor full documentation.See also
numpy.cumsumequivalent function
-
decompose(self, bases=[])¶ Generates a new
Quantitywith the units decomposed. Decomposed units have only irreducible units in them (seeastropy.units.UnitBase.decompose).- Parameters
bases : sequence of UnitBase, optional
The bases to decompose into. When not provided, decomposes down to any irreducible units. When provided, the decomposed result will only contain the given units. This will raises a
UnitsErrorif it’s not possible to do so.- Returns
newq :
QuantityA new object equal to this quantity with units decomposed.
-
diagonal(offset=0, axis1=0, axis2=1)¶ Return specified diagonals. In NumPy 1.9 the returned array is a read-only view instead of a copy as in previous NumPy versions. In a future version the read-only restriction will be removed.
Refer to
numpy.diagonal()for full documentation.See also
numpy.diagonalequivalent function
-
diff(self, n=1, axis=-1)[source]¶ Calculate the n-th order discrete difference along given axis.
The first order difference is given by
out[n] = a[n+1] - a[n]along the given axis, higher order differences are calculated by usingdiffrecursively.- Parameters
n : int, optional
The number of times values are differenced.
axis : int, optional
The axis along which the difference is taken, default is the last axis.
- Returns
diff :
SeriesThe
norder differences. The shape of the output is the same as the input, except alongaxiswhere the dimension is smaller byn.
See also
numpy.difffor documentation on the underlying method
-
dot(b, out=None)¶ Dot product of two arrays.
Refer to
numpy.dotfor full documentation.See also
numpy.dotequivalent function
Examples
>>> a = np.eye(2) >>> b = np.ones((2, 2)) * 2 >>> a.dot(b) array([[2., 2.], [2., 2.]])
This array method can be conveniently chained:
>>> a.dot(b).dot(b) array([[8., 8.], [8., 8.]])
-
dump(file)¶ Dump a pickle of the array to the specified file. The array can be read back with pickle.load or numpy.load.
- Parameters
file : str or Path
A string naming the dump file.
Changed in version 1.17.0:
pathlib.Pathobjects are now accepted.
-
dumps()[source]¶ Returns the pickle of the array as a string. pickle.loads or numpy.loads will convert the string back to an array.
- Parameters
- None
-
ediff1d(self, to_end=None, to_begin=None)¶
-
fill(value)¶ Fill the array with a scalar value.
- Parameters
value : scalar
All elements of
awill be assigned this value.
Examples
>>> a = np.array([1, 2]) >>> a.fill(0) >>> a array([0, 0]) >>> a = np.empty(2) >>> a.fill(1) >>> a array([1., 1.])
-
filter(self, *filt, **kwargs)[source]¶ Apply the given filter to this
Spectrogram.- Parameters
*filt : filter arguments
analog :
bool, optionalinplace :
bool, optional- Returns
result :
Spectrogramthe filtered version of the input
Spectrogram, ifinplace=Truewas given, this is just a reference to the modified input array- Raises
ValueError
if
filtarguments cannot be interpreted properly
-
flatten(self, order='C')[source]¶ Return a copy of the array collapsed into one dimension.
Any index information is removed as part of the flattening, and the result is returned as a
Quantityarray.- Parameters
order : {‘C’, ‘F’, ‘A’, ‘K’}, optional
‘C’ means to flatten in row-major (C-style) order. ‘F’ means to flatten in column-major (Fortran- style) order. ‘A’ means to flatten in column-major order if
ais Fortran contiguous in memory, row-major order otherwise. ‘K’ means to flattenain the order the elements occur in memory. The default is ‘C’.- Returns
y :
QuantityA copy of the input array, flattened to one dimension.
Examples
>>> a = Array([[1,2], [3,4]], unit='m', name='Test') >>> a.flatten() <Quantity [1., 2., 3., 4.] m>
-
classmethod
from_spectra(*spectra, **kwargs)[source]¶ Build a new
Spectrogramfrom a list of spectra.- Parameters
*spectra
any number of
FrequencySeriesseriesdt :
float,Quantity, optionalstride between given spectra
- Returns
Spectrogram
a new
Spectrogramfrom a vertical stacking of the spectra The new object takes the metadata from the first givenFrequencySeriesif not given explicitly
Notes
Each
FrequencySeriespassed to this constructor must be the same length.
-
getfield(dtype, offset=0)¶ Returns a field of the given array as a certain type.
A field is a view of the array data with a given data-type. The values in the view are determined by the given type and the offset into the current array in bytes. The offset needs to be such that the view dtype fits in the array dtype; for example an array of dtype complex128 has 16-byte elements. If taking a view with a 32-bit integer (4 bytes), the offset needs to be between 0 and 12 bytes.
- Parameters
dtype : str or dtype
The data type of the view. The dtype size of the view can not be larger than that of the array itself.
offset : int
Number of bytes to skip before beginning the element view.
Examples
>>> x = np.diag([1.+1.j]*2) >>> x[1, 1] = 2 + 4.j >>> x array([[1.+1.j, 0.+0.j], [0.+0.j, 2.+4.j]]) >>> x.getfield(np.float64) array([[1., 0.], [0., 2.]])
By choosing an offset of 8 bytes we can select the complex part of the array for our view:
>>> x.getfield(np.float64, offset=8) array([[1., 0.], [0., 4.]])
-
inject(self, other)[source]¶ Add two compatible
Seriesalong their shared x-axis values.- Parameters
other :
Seriesa
Serieswhose xindex intersects withself.xindex- Returns
out :
Seriesthe sum of
selfandotheralong their shared x-axis values- Raises
ValueError
if
selfandotherhave incompatible units or xindex intervals
Notes
If
other.xindexandself.xindexdo not intersect, this method will return a copy ofself. If the series have uniformly offset indices, this method will raise a warning.If
self.xindexis an array of timestamps, and ifother.xspanis not a subset ofself.xspan, thenotherwill be cropped before being adding toself.Users who wish to taper or window their
Seriesshould do so before passing it to this method. SeeTimeSeries.taper()andplanck()for more information.
-
insert(self, obj, values, axis=None)¶ Insert values along the given axis before the given indices and return a new
Quantityobject.This is a thin wrapper around the
numpy.insertfunction.- Parameters
obj : int, slice or sequence of ints
Object that defines the index or indices before which
valuesis inserted.values : array-like
Values to insert. If the type of
valuesis different from that of quantity,valuesis converted to the matching type.valuesshould be shaped so that it can be broadcast appropriately The unit ofvaluesmust be consistent with this quantity.axis : int, optional
Axis along which to insert
values. Ifaxisis None then the quantity array is flattened before insertion.- Returns
out :
QuantityA copy of quantity with
valuesinserted. Note that the insertion does not occur in-place: a new quantity array is returned.
Examples
>>> import astropy.units as u >>> q = [1, 2] * u.m >>> q.insert(0, 50 * u.cm) <Quantity [ 0.5, 1., 2.] m>
>>> q = [[1, 2], [3, 4]] * u.m >>> q.insert(1, [10, 20] * u.m, axis=0) <Quantity [[ 1., 2.], [ 10., 20.], [ 3., 4.]] m>
>>> q.insert(1, 10 * u.m, axis=1) <Quantity [[ 1., 10., 2.], [ 3., 10., 4.]] m>
-
is_contiguous(self, other, tol=3.814697265625e-06)[source]¶ Check whether other is contiguous with self.
- Parameters
other :
Series,numpy.ndarrayanother series of the same type to test for contiguity
tol :
float, optionalthe numerical tolerance of the test
- Returns
1
if
otheris contiguous with this series, i.e. would attach seamlessly onto the end- -1
if
otheris anti-contiguous with this seires, i.e. would attach seamlessly onto the start
0
if
otheris completely dis-contiguous with thie series
Notes
if a raw
numpy.ndarrayis passed as other, with no metadata, then the contiguity check will always pass
-
item(*args)¶ Copy an element of an array to a standard Python scalar and return it.
- Parameters
*args : Arguments (variable number and type)
none: in this case, the method only works for arrays with one element (
a.size == 1), which element is copied into a standard Python scalar object and returned.int_type: this argument is interpreted as a flat index into the array, specifying which element to copy and return.
tuple of int_types: functions as does a single int_type argument, except that the argument is interpreted as an nd-index into the array.
- Returns
z : Standard Python scalar object
A copy of the specified element of the array as a suitable Python scalar
Notes
When the data type of
ais longdouble or clongdouble, item() returns a scalar array object because there is no available Python scalar that would not lose information. Void arrays return a buffer object for item(), unless fields are defined, in which case a tuple is returned.itemis very similar to a[args], except, instead of an array scalar, a standard Python scalar is returned. This can be useful for speeding up access to elements of the array and doing arithmetic on elements of the array using Python’s optimized math.Examples
>>> np.random.seed(123) >>> x = np.random.randint(9, size=(3, 3)) >>> x array([[2, 2, 6], [1, 3, 6], [1, 0, 1]]) >>> x.item(3) 1 >>> x.item(7) 0 >>> x.item((0, 1)) 2 >>> x.item((2, 2)) 1
-
itemset(*args)¶ Insert scalar into an array (scalar is cast to array’s dtype, if possible)
There must be at least 1 argument, and define the last argument as item. Then,
a.itemset(*args)is equivalent to but faster thana[args] = item. The item should be a scalar value andargsmust select a single item in the arraya.- Parameters
*args : Arguments
If one argument: a scalar, only used in case
ais of size 1. If two arguments: the last argument is the value to be set and must be a scalar, the first argument specifies a single array element location. It is either an int or a tuple.
Notes
Compared to indexing syntax,
itemsetprovides some speed increase for placing a scalar into a particular location in anndarray, if you must do this. However, generally this is discouraged: among other problems, it complicates the appearance of the code. Also, when usingitemset(anditem) inside a loop, be sure to assign the methods to a local variable to avoid the attribute look-up at each loop iteration.Examples
>>> np.random.seed(123) >>> x = np.random.randint(9, size=(3, 3)) >>> x array([[2, 2, 6], [1, 3, 6], [1, 0, 1]]) >>> x.itemset(4, 0) >>> x.itemset((2, 2), 9) >>> x array([[2, 2, 6], [1, 0, 6], [1, 0, 9]])
-
max(axis=None, out=None, keepdims=False, initial=<no value>, where=True)¶ Return the maximum along a given axis.
Refer to
numpy.amaxfor full documentation.See also
numpy.amaxequivalent function
-
mean(axis=None, dtype=None, out=None, keepdims=False)¶ Returns the average of the array elements along given axis.
Refer to
numpy.meanfor full documentation.See also
numpy.meanequivalent function
-
median(self, axis=None, **kwargs)[source]¶ Compute the median along the specified axis.
Returns the median of the array elements.
- Parameters
a : array_like
Input array or object that can be converted to an array.
axis : {int, sequence of int, None}, optional
Axis or axes along which the medians are computed. The default is to compute the median along a flattened version of the array. A sequence of axes is supported since version 1.9.0.
out : ndarray, optional
Alternative output array in which to place the result. It must have the same shape and buffer length as the expected output, but the type (of the output) will be cast if necessary.
overwrite_input : bool, optional
If True, then allow use of memory of input array
afor calculations. The input array will be modified by the call tomedian. This will save memory when you do not need to preserve the contents of the input array. Treat the input as undefined, but it will probably be fully or partially sorted. Default is False. Ifoverwrite_inputisTrueandais not already anndarray, an error will be raised.keepdims : bool, optional
If this is set to True, the axes which are reduced are left in the result as dimensions with size one. With this option, the result will broadcast correctly against the original
arr.New in version 1.9.0.
- Returns
median : ndarray
A new array holding the result. If the input contains integers or floats smaller than
float64, then the output data-type isnp.float64. Otherwise, the data-type of the output is the same as that of the input. Ifoutis specified, that array is returned instead.
See also
Notes
Given a vector
Vof lengthN, the median ofVis the middle value of a sorted copy ofV,V_sorted- i e.,V_sorted[(N-1)/2], whenNis odd, and the average of the two middle values ofV_sortedwhenNis even.Examples
>>> a = np.array([[10, 7, 4], [3, 2, 1]]) >>> a array([[10, 7, 4], [ 3, 2, 1]]) >>> np.median(a) 3.5 >>> np.median(a, axis=0) array([6.5, 4.5, 2.5]) >>> np.median(a, axis=1) array([7., 2.]) >>> m = np.median(a, axis=0) >>> out = np.zeros_like(m) >>> np.median(a, axis=0, out=m) array([6.5, 4.5, 2.5]) >>> m array([6.5, 4.5, 2.5]) >>> b = a.copy() >>> np.median(b, axis=1, overwrite_input=True) array([7., 2.]) >>> assert not np.all(a==b) >>> b = a.copy() >>> np.median(b, axis=None, overwrite_input=True) 3.5 >>> assert not np.all(a==b)
-
min(axis=None, out=None, keepdims=False, initial=<no value>, where=True)¶ Return the minimum along a given axis.
Refer to
numpy.aminfor full documentation.See also
numpy.aminequivalent function
-
nansum(self, axis=None, out=None, keepdims=False)¶
-
newbyteorder(new_order='S')¶ Return the array with the same data viewed with a different byte order.
Equivalent to:
arr.view(arr.dtype.newbytorder(new_order))
Changes are also made in all fields and sub-arrays of the array data type.
- Parameters
new_order : string, optional
Byte order to force; a value from the byte order specifications below.
new_ordercodes can be any of:‘S’ - swap dtype from current to opposite endian
{‘<’, ‘L’} - little endian
{‘>’, ‘B’} - big endian
{‘=’, ‘N’} - native order
{‘|’, ‘I’} - ignore (no change to byte order)
The default value (‘S’) results in swapping the current byte order. The code does a case-insensitive check on the first letter of
new_orderfor the alternatives above. For example, any of ‘B’ or ‘b’ or ‘biggish’ are valid to specify big-endian.- Returns
new_arr : array
New array object with the dtype reflecting given change to the byte order.
-
nonzero()¶ Return the indices of the elements that are non-zero.
Refer to
numpy.nonzerofor full documentation.See also
numpy.nonzeroequivalent function
-
override_unit(self, unit, parse_strict='raise')[source]¶ Forcefully reset the unit of these data
Use of this method is discouraged in favour of
to(), which performs accurate conversions from one unit to another. The method should really only be used when the original unit of the array is plain wrong.- Parameters
-
the unit to force onto this array
parse_strict :
str, optionalhow to handle errors in the unit parsing, default is to raise the underlying exception from
astropy.units - Raises
ValueError
if a
strcannot be parsed as a valid unit
-
pad(self, pad_width, **kwargs)[source]¶ Pad this series to a new size
- Parameters
pad_width :
int, pair ofintsnumber of samples by which to pad each end of the array. Single int to pad both ends by the same amount, or (before, after)
tupleto give uneven padding**kwargs
see
numpy.pad()for kwarg documentation- Returns
series :
Seriesthe padded version of the input
See also
numpy.padfor details on the underlying functionality
-
partition(kth, axis=-1, kind='introselect', order=None)¶ Rearranges the elements in the array in such a way that the value of the element in kth position is in the position it would be in a sorted array. All elements smaller than the kth element are moved before this element and all equal or greater are moved behind it. The ordering of the elements in the two partitions is undefined.
New in version 1.8.0.
- Parameters
kth : int or sequence of ints
Element index to partition by. The kth element value will be in its final sorted position and all smaller elements will be moved before it and all equal or greater elements behind it. The order of all elements in the partitions is undefined. If provided with a sequence of kth it will partition all elements indexed by kth of them into their sorted position at once.
axis : int, optional
Axis along which to sort. Default is -1, which means sort along the last axis.
kind : {‘introselect’}, optional
Selection algorithm. Default is ‘introselect’.
order : str or list of str, optional
When
ais an array with fields defined, this argument specifies which fields to compare first, second, etc. A single field can be specified as a string, and not all fields need to be specified, but unspecified fields will still be used, in the order in which they come up in the dtype, to break ties.
See also
numpy.partitionReturn a parititioned copy of an array.
argpartitionIndirect partition.
sortFull sort.
Notes
See
np.partitionfor notes on the different algorithms.Examples
>>> a = np.array([3, 4, 2, 1]) >>> a.partition(3) >>> a array([2, 1, 3, 4])
>>> a.partition((1, 3)) >>> a array([1, 2, 3, 4])
-
percentile(self, percentile)[source]¶ Calculate a given spectral percentile for this
Spectrogram.- Parameters
percentile :
floatpercentile (0 - 100) of the bins to compute
- Returns
spectrum :
FrequencySeriesthe given percentile
FrequencySeriescalculated from thisSpectralVaraicence
-
plot(self, figsize=(12, 6), xscale='auto-gps', **kwargs)[source]¶ Plot the data for this
Spectrogram- Parameters
**kwargs
all keyword arguments are passed along to underlying functions, see below for references
- Returns
plot :
Plotthe
Plotcontaining the data
See also
matplotlib.pyplot.figurefor documentation of keyword arguments used to create the figure
matplotlib.figure.Figure.add_subplotfor documentation of keyword arguments used to create the axes
gwpy.plot.Axes.imshowgwpy.plot.Axes.pcolormeshfor documentation of keyword arguments used in rendering the
Spectrogramdata
-
prepend(self, other, inplace=True, pad=None, gap=None, resize=True)[source]¶ Connect another series onto the start of the current one.
- Parameters
other :
Seriesanother series of the same type as this one
inplace :
bool, optionalperform operation in-place, modifying current series, otherwise copy data and return new series, default:
TrueWarning
inplaceprepend bypasses the reference check innumpy.ndarray.resize, so be carefully to only use this for arrays that haven’t been sharing their memory!pad :
float, optionalvalue with which to pad discontiguous series, by default gaps will result in a
ValueError.gap :
str, optionalaction to perform if there’s a gap between the other series and this one. One of
'raise'- raise aValueError'ignore'- remove gap and join data'pad'- pad gap with zeros
If
padis given and is notNone, the default is'pad', otherwise'raise'.resize :
bool, optionalresize this array to accommodate new data, otherwise shift the old data to the left (potentially falling off the start) and put the new data in at the end, default:
True.- Returns
series :
TimeSeriestime-series containing joined data sets
-
prod(axis=None, dtype=None, out=None, keepdims=False, initial=1, where=True)¶ Return the product of the array elements over the given axis
Refer to
numpy.prodfor full documentation.See also
numpy.prodequivalent function
-
ptp(axis=None, out=None, keepdims=False)¶ Peak to peak (maximum - minimum) value along a given axis.
Refer to
numpy.ptpfor full documentation.See also
numpy.ptpequivalent function
-
put(indices, values, mode='raise')¶ Set
a.flat[n] = values[n]for allnin indices.Refer to
numpy.putfor full documentation.See also
numpy.putequivalent function
-
ratio(self, operand)[source]¶ Calculate the ratio of this
Spectrogramagainst a reference- Parameters
operand :
str,FrequencySeries,Quantitya
FrequencySeriesorQuantityto weight against, or one of'mean': weight against the mean of each spectrum in this Spectrogram'median': weight against the median of each spectrum in this Spectrogram
- Returns
spectrogram :
Spectrograma new
Spectrogram- Raises
ValueError
if
operandis given as astrthat isn’t supported
-
ravel([order])¶ Return a flattened array.
Refer to
numpy.ravelfor full documentation.See also
numpy.ravelequivalent function
ndarray.flata flat iterator on the array.
-
classmethod
read(source, *args, **kwargs)[source]¶ Read data into a
SpectrogramArguments and keywords depend on the output format, see the online documentation for full details for each format, the parameters below are common to most formats.
- Parameters
-
*args
Other arguments are (in general) specific to the given
format.format :
str, optionalSource format identifier. If not given, the format will be detected if possible. See below for list of acceptable formats.
**kwargs
Other keywords are (in general) specific to the given
format. - Returns
- specgram
Spectrogram
- specgram
Notes
The available built-in formats are:
Format
Read
Write
Auto-identify
hdf5
Yes
No
No
-
repeat(repeats, axis=None)¶ Repeat elements of an array.
Refer to
numpy.repeatfor full documentation.See also
numpy.repeatequivalent function
-
reshape(shape, order='C')¶ Returns an array containing the same data with a new shape.
Refer to
numpy.reshapefor full documentation.See also
numpy.reshapeequivalent function
Notes
Unlike the free function
numpy.reshape, this method onndarrayallows the elements of the shape parameter to be passed in as separate arguments. For example,a.reshape(10, 11)is equivalent toa.reshape((10, 11)).
-
resize(new_shape, refcheck=True)¶ Change shape and size of array in-place.
- Parameters
new_shape : tuple of ints, or
nintsShape of resized array.
refcheck : bool, optional
If False, reference count will not be checked. Default is True.
- Returns
- None
- Raises
ValueError
If
adoes not own its own data or references or views to it exist, and the data memory must be changed. PyPy only: will always raise if the data memory must be changed, since there is no reliable way to determine if references or views to it exist.SystemError
If the
orderkeyword argument is specified. This behaviour is a bug in NumPy.
See also
resizeReturn a new array with the specified shape.
Notes
This reallocates space for the data area if necessary.
Only contiguous arrays (data elements consecutive in memory) can be resized.
The purpose of the reference count check is to make sure you do not use this array as a buffer for another Python object and then reallocate the memory. However, reference counts can increase in other ways so if you are sure that you have not shared the memory for this array with another Python object, then you may safely set
refcheckto False.Examples
Shrinking an array: array is flattened (in the order that the data are stored in memory), resized, and reshaped:
>>> a = np.array([[0, 1], [2, 3]], order='C') >>> a.resize((2, 1)) >>> a array([[0], [1]])
>>> a = np.array([[0, 1], [2, 3]], order='F') >>> a.resize((2, 1)) >>> a array([[0], [2]])
Enlarging an array: as above, but missing entries are filled with zeros:
>>> b = np.array([[0, 1], [2, 3]]) >>> b.resize(2, 3) # new_shape parameter doesn't have to be a tuple >>> b array([[0, 1, 2], [3, 0, 0]])
Referencing an array prevents resizing…
>>> c = a >>> a.resize((1, 1)) Traceback (most recent call last): ... ValueError: cannot resize an array that references or is referenced ...
Unless
refcheckis False:>>> a.resize((1, 1), refcheck=False) >>> a array([[0]]) >>> c array([[0]])
-
round(decimals=0, out=None)¶ Return
awith each element rounded to the given number of decimals.Refer to
numpy.aroundfor full documentation.See also
numpy.aroundequivalent function
-
searchsorted(v, side='left', sorter=None)¶ Find indices where elements of v should be inserted in a to maintain order.
For full documentation, see
numpy.searchsortedSee also
numpy.searchsortedequivalent function
-
setfield(val, dtype, offset=0)¶ Put a value into a specified place in a field defined by a data-type.
Place
valintoa’s field defined bydtypeand beginningoffsetbytes into the field.- Parameters
val : object
Value to be placed in field.
dtype : dtype object
Data-type of the field in which to place
val.offset : int, optional
The number of bytes into the field at which to place
val.- Returns
- None
See also
Examples
>>> x = np.eye(3) >>> x.getfield(np.float64) array([[1., 0., 0.], [0., 1., 0.], [0., 0., 1.]]) >>> x.setfield(3, np.int32) >>> x.getfield(np.int32) array([[3, 3, 3], [3, 3, 3], [3, 3, 3]], dtype=int32) >>> x array([[1.0e+000, 1.5e-323, 1.5e-323], [1.5e-323, 1.0e+000, 1.5e-323], [1.5e-323, 1.5e-323, 1.0e+000]]) >>> x.setfield(np.eye(3), np.int32) >>> x array([[1., 0., 0.], [0., 1., 0.], [0., 0., 1.]])
-
setflags(write=None, align=None, uic=None)¶ Set array flags WRITEABLE, ALIGNED, (WRITEBACKIFCOPY and UPDATEIFCOPY), respectively.
These Boolean-valued flags affect how numpy interprets the memory area used by
a(see Notes below). The ALIGNED flag can only be set to True if the data is actually aligned according to the type. The WRITEBACKIFCOPY and (deprecated) UPDATEIFCOPY flags can never be set to True. The flag WRITEABLE can only be set to True if the array owns its own memory, or the ultimate owner of the memory exposes a writeable buffer interface, or is a string. (The exception for string is made so that unpickling can be done without copying memory.)- Parameters
write : bool, optional
Describes whether or not
acan be written to.align : bool, optional
Describes whether or not
ais aligned properly for its type.uic : bool, optional
Describes whether or not
ais a copy of another “base” array.
Notes
Array flags provide information about how the memory area used for the array is to be interpreted. There are 7 Boolean flags in use, only four of which can be changed by the user: WRITEBACKIFCOPY, UPDATEIFCOPY, WRITEABLE, and ALIGNED.
WRITEABLE (W) the data area can be written to;
ALIGNED (A) the data and strides are aligned appropriately for the hardware (as determined by the compiler);
UPDATEIFCOPY (U) (deprecated), replaced by WRITEBACKIFCOPY;
WRITEBACKIFCOPY (X) this array is a copy of some other array (referenced by .base). When the C-API function PyArray_ResolveWritebackIfCopy is called, the base array will be updated with the contents of this array.
All flags can be accessed using the single (upper case) letter as well as the full name.
Examples
>>> y = np.array([[3, 1, 7], ... [2, 0, 0], ... [8, 5, 9]]) >>> y array([[3, 1, 7], [2, 0, 0], [8, 5, 9]]) >>> y.flags C_CONTIGUOUS : True F_CONTIGUOUS : False OWNDATA : True WRITEABLE : True ALIGNED : True WRITEBACKIFCOPY : False UPDATEIFCOPY : False >>> y.setflags(write=0, align=0) >>> y.flags C_CONTIGUOUS : True F_CONTIGUOUS : False OWNDATA : True WRITEABLE : False ALIGNED : False WRITEBACKIFCOPY : False UPDATEIFCOPY : False >>> y.setflags(uic=1) Traceback (most recent call last): File "<stdin>", line 1, in <module> ValueError: cannot set WRITEBACKIFCOPY flag to True
-
shift(self, delta)[source]¶ Shift this
Seriesforward on the X-axis bydeltaThis modifies the series in-place.
- Parameters
-
The amount by which to shift (in x-axis units if
float), give a negative value to shift backwards in time
Examples
>>> from gwpy.types import Series >>> a = Series([1, 2, 3, 4, 5], x0=0, dx=1, xunit='m') >>> print(a.x0) 0.0 m >>> a.shift(5) >>> print(a.x0) 5.0 m >>> a.shift('-1 km') -995.0 m
-
sort(axis=-1, kind=None, order=None)¶ Sort an array in-place. Refer to
numpy.sortfor full documentation.- Parameters
axis : int, optional
Axis along which to sort. Default is -1, which means sort along the last axis.
kind : {‘quicksort’, ‘mergesort’, ‘heapsort’, ‘stable’}, optional
Sorting algorithm. The default is ‘quicksort’. Note that both ‘stable’ and ‘mergesort’ use timsort under the covers and, in general, the actual implementation will vary with datatype. The ‘mergesort’ option is retained for backwards compatibility.
Changed in version 1.15.0.: The ‘stable’ option was added.
order : str or list of str, optional
When
ais an array with fields defined, this argument specifies which fields to compare first, second, etc. A single field can be specified as a string, and not all fields need be specified, but unspecified fields will still be used, in the order in which they come up in the dtype, to break ties.
See also
numpy.sortReturn a sorted copy of an array.
argsortIndirect sort.
lexsortIndirect stable sort on multiple keys.
searchsortedFind elements in sorted array.
partitionPartial sort.
Notes
See
numpy.sortfor notes on the different sorting algorithms.Examples
>>> a = np.array([[1,4], [3,1]]) >>> a.sort(axis=1) >>> a array([[1, 4], [1, 3]]) >>> a.sort(axis=0) >>> a array([[1, 3], [1, 4]])
Use the
orderkeyword to specify a field to use when sorting a structured array:>>> a = np.array([('a', 2), ('c', 1)], dtype=[('x', 'S1'), ('y', int)]) >>> a.sort(order='y') >>> a array([(b'c', 1), (b'a', 2)], dtype=[('x', 'S1'), ('y', '<i8')])
-
squeeze(axis=None)¶ Remove single-dimensional entries from the shape of
a.Refer to
numpy.squeezefor full documentation.See also
numpy.squeezeequivalent function
-
std(axis=None, dtype=None, out=None, ddof=0, keepdims=False)¶ Returns the standard deviation of the array elements along given axis.
Refer to
numpy.stdfor full documentation.See also
numpy.stdequivalent function
-
sum(axis=None, dtype=None, out=None, keepdims=False, initial=0, where=True)¶ Return the sum of the array elements over the given axis.
Refer to
numpy.sumfor full documentation.See also
numpy.sumequivalent function
-
swapaxes(axis1, axis2)¶ Return a view of the array with
axis1andaxis2interchanged.Refer to
numpy.swapaxesfor full documentation.See also
numpy.swapaxesequivalent function
-
take(indices, axis=None, out=None, mode='raise')¶ Return an array formed from the elements of
aat the given indices.Refer to
numpy.takefor full documentation.See also
numpy.takeequivalent function
-
to(self, unit, equivalencies=[])¶ Return a new
Quantityobject with the specified unit.- Parameters
unit :
UnitBaseinstance, strequivalencies : list of equivalence pairs, optional
A list of equivalence pairs to try if the units are not directly convertible. See Equivalencies. If not provided or
[], class default equivalencies will be used (none forQuantity, but may be set for subclasses) IfNone, no equivalencies will be applied at all, not even any set globally or within a context.
See also
to_valueget the numerical value in a given unit.
-
to_string(self, unit=None, precision=None, format=None, subfmt=None)¶ Generate a string representation of the quantity and its unit.
The behavior of this function can be altered via the
numpy.set_printoptionsfunction and its various keywords. The exception to this is thethresholdkeyword, which is controlled via the[units.quantity]configuration itemlatex_array_threshold. This is treated separately because the numpy default of 1000 is too big for most browsers to handle.- Parameters
unit :
UnitBase, optionalSpecifies the unit. If not provided, the unit used to initialize the quantity will be used.
precision : numeric, optional
The level of decimal precision. If
None, or not provided, it will be determined from NumPy print options.format : str, optional
The format of the result. If not provided, an unadorned string is returned. Supported values are:
‘latex’: Return a LaTeX-formatted string
subfmt : str, optional
Subformat of the result. For the moment, only used for format=”latex”. Supported values are:
‘inline’: Use
$ ... $as delimiters.‘display’: Use
$\displaystyle ... $as delimiters.
- Returns
lstr
A string with the contents of this Quantity
-
to_value(self, unit=None, equivalencies=[])¶ The numerical value, possibly in a different unit.
- Parameters
unit :
UnitBaseinstance or str, optionalThe unit in which the value should be given. If not given or
None, use the current unit.equivalencies : list of equivalence pairs, optional
A list of equivalence pairs to try if the units are not directly convertible (see Equivalencies). If not provided or
[], class default equivalencies will be used (none forQuantity, but may be set for subclasses). IfNone, no equivalencies will be applied at all, not even any set globally or within a context.- Returns
value :
ndarrayor scalarThe value in the units specified. For arrays, this will be a view of the data if no unit conversion was necessary.
See also
toGet a new instance in a different unit.
-
tobytes(order='C')¶ Construct Python bytes containing the raw data bytes in the array.
Constructs Python bytes showing a copy of the raw contents of data memory. The bytes object can be produced in either ‘C’ or ‘Fortran’, or ‘Any’ order (the default is ‘C’-order). ‘Any’ order means C-order unless the F_CONTIGUOUS flag in the array is set, in which case it means ‘Fortran’ order.
New in version 1.9.0.
- Parameters
order : {‘C’, ‘F’, None}, optional
Order of the data for multidimensional arrays: C, Fortran, or the same as for the original array.
- Returns
s : bytes
Python bytes exhibiting a copy of
a’s raw data.
Examples
>>> x = np.array([[0, 1], [2, 3]], dtype='<u2') >>> x.tobytes() b'\x00\x00\x01\x00\x02\x00\x03\x00' >>> x.tobytes('C') == x.tobytes() True >>> x.tobytes('F') b'\x00\x00\x02\x00\x01\x00\x03\x00'
-
tofile(fid, sep="", format="%s")¶ Write array to a file as text or binary (default).
Data is always written in ‘C’ order, independent of the order of
a. The data produced by this method can be recovered using the function fromfile().- Parameters
fid : file or str or Path
An open file object, or a string containing a filename.
Changed in version 1.17.0:
pathlib.Pathobjects are now accepted.sep : str
Separator between array items for text output. If “” (empty), a binary file is written, equivalent to
file.write(a.tobytes()).format : str
Format string for text file output. Each entry in the array is formatted to text by first converting it to the closest Python type, and then using “format” % item.
Notes
This is a convenience function for quick storage of array data. Information on endianness and precision is lost, so this method is not a good choice for files intended to archive data or transport data between machines with different endianness. Some of these problems can be overcome by outputting the data as text files, at the expense of speed and file size.
When fid is a file object, array contents are directly written to the file, bypassing the file object’s
writemethod. As a result, tofile cannot be used with files objects supporting compression (e.g., GzipFile) or file-like objects that do not supportfileno()(e.g., BytesIO).
-
tolist()¶ Return the array as an
a.ndim-levels deep nested list of Python scalars.Return a copy of the array data as a (nested) Python list. Data items are converted to the nearest compatible builtin Python type, via the
itemfunction.If
a.ndimis 0, then since the depth of the nested list is 0, it will not be a list at all, but a simple Python scalar.- Parameters
- none
- Returns
y : object, or list of object, or list of list of object, or …
The possibly nested list of array elements.
Notes
The array may be recreated via
a = np.array(a.tolist()), although this may sometimes lose precision.Examples
For a 1D array,
a.tolist()is almost the same aslist(a):>>> a = np.array([1, 2]) >>> list(a) [1, 2] >>> a.tolist() [1, 2]
However, for a 2D array,
tolistapplies recursively:>>> a = np.array([[1, 2], [3, 4]]) >>> list(a) [array([1, 2]), array([3, 4])] >>> a.tolist() [[1, 2], [3, 4]]
The base case for this recursion is a 0D array:
>>> a = np.array(1) >>> list(a) Traceback (most recent call last): ... TypeError: iteration over a 0-d array >>> a.tolist() 1
-
tostring(order='C')[source]¶ Construct Python bytes containing the raw data bytes in the array.
Constructs Python bytes showing a copy of the raw contents of data memory. The bytes object can be produced in either ‘C’ or ‘Fortran’, or ‘Any’ order (the default is ‘C’-order). ‘Any’ order means C-order unless the F_CONTIGUOUS flag in the array is set, in which case it means ‘Fortran’ order.
This function is a compatibility alias for tobytes. Despite its name it returns bytes not strings.
- Parameters
order : {‘C’, ‘F’, None}, optional
Order of the data for multidimensional arrays: C, Fortran, or the same as for the original array.
- Returns
s : bytes
Python bytes exhibiting a copy of
a’s raw data.
Examples
>>> x = np.array([[0, 1], [2, 3]], dtype='<u2') >>> x.tobytes() b'\x00\x00\x01\x00\x02\x00\x03\x00' >>> x.tobytes('C') == x.tobytes() True >>> x.tobytes('F') b'\x00\x00\x02\x00\x01\x00\x03\x00'
-
trace(offset=0, axis1=0, axis2=1, dtype=None, out=None)¶ Return the sum along diagonals of the array.
Refer to
numpy.tracefor full documentation.See also
numpy.traceequivalent function
-
transpose(*axes)¶ Returns a view of the array with axes transposed.
For a 1-D array this has no effect, as a transposed vector is simply the same vector. To convert a 1-D array into a 2D column vector, an additional dimension must be added.
np.atleast2d(a).Tachieves this, as doesa[:, np.newaxis]. For a 2-D array, this is a standard matrix transpose. For an n-D array, if axes are given, their order indicates how the axes are permuted (see Examples). If axes are not provided anda.shape = (i[0], i[1], ... i[n-2], i[n-1]), thena.transpose().shape = (i[n-1], i[n-2], ... i[1], i[0]).- Parameters
axes : None, tuple of ints, or
nintsNone or no argument: reverses the order of the axes.
tuple of ints:
iin thej-th place in the tuple meansa’si-th axis becomesa.transpose()’sj-th axis.nints: same as an n-tuple of the same ints (this form is intended simply as a “convenience” alternative to the tuple form)
- Returns
out : ndarray
View of
a, with axes suitably permuted.
See also
ndarray.TArray property returning the array transposed.
ndarray.reshapeGive a new shape to an array without changing its data.
Examples
>>> a = np.array([[1, 2], [3, 4]]) >>> a array([[1, 2], [3, 4]]) >>> a.transpose() array([[1, 3], [2, 4]]) >>> a.transpose((1, 0)) array([[1, 3], [2, 4]]) >>> a.transpose(1, 0) array([[1, 3], [2, 4]])
-
update(self, other, inplace=True)[source]¶ Update this series by appending new data from an other and dropping the same amount of data off the start.
This is a convenience method that just calls
appendwithresize=False.
-
var(axis=None, dtype=None, out=None, ddof=0, keepdims=False)¶ Returns the variance of the array elements, along given axis.
Refer to
numpy.varfor full documentation.See also
numpy.varequivalent function
-
variance(self, bins=None, low=None, high=None, nbins=500, log=False, norm=False, density=False)[source]¶ Calculate the
SpectralVarianceof thisSpectrogram.- Parameters
bins :
ndarray, optional, defaultNonearray of histogram bin edges, including the rightmost edge
low :
float, optional, default:Noneleft edge of lowest amplitude bin, only read if
binsis not givenhigh :
float, optional, default:Noneright edge of highest amplitude bin, only read if
binsis not givennbins :
int, optional, default:500number of bins to generate, only read if
binsis not givenlog :
bool, optional, default:Falsecalculate amplitude bins over a logarithmic scale, only read if
binsis not givennorm :
bool, optional, default:Falsenormalise bin counts to a unit sum
density :
bool, optional, default:Falsenormalise bin counts to a unit integral
- Returns
specvar :
SpectralVariance2D-array of spectral frequency-amplitude counts
See also
numpy.histogramfor details on specifying bins and weights
-
view(dtype=None, type=None)¶ New view of array with the same data.
- Parameters
dtype : data-type or ndarray sub-class, optional
Data-type descriptor of the returned view, e.g., float32 or int16. The default, None, results in the view having the same data-type as
a. This argument can also be specified as an ndarray sub-class, which then specifies the type of the returned object (this is equivalent to setting thetypeparameter).type : Python type, optional
Type of the returned view, e.g., ndarray or matrix. Again, the default None results in type preservation.
Notes
a.view()is used two different ways:a.view(some_dtype)ora.view(dtype=some_dtype)constructs a view of the array’s memory with a different data-type. This can cause a reinterpretation of the bytes of memory.a.view(ndarray_subclass)ora.view(type=ndarray_subclass)just returns an instance ofndarray_subclassthat looks at the same array (same shape, dtype, etc.) This does not cause a reinterpretation of the memory.For
a.view(some_dtype), ifsome_dtypehas a different number of bytes per entry than the previous dtype (for example, converting a regular array to a structured array), then the behavior of the view cannot be predicted just from the superficial appearance ofa(shown byprint(a)). It also depends on exactly howais stored in memory. Therefore ifais C-ordered versus fortran-ordered, versus defined as a slice or transpose, etc., the view may give different results.Examples
>>> x = np.array([(1, 2)], dtype=[('a', np.int8), ('b', np.int8)])
Viewing array data using a different type and dtype:
>>> y = x.view(dtype=np.int16, type=np.matrix) >>> y matrix([[513]], dtype=int16) >>> print(type(y)) <class 'numpy.matrix'>
Creating a view on a structured array so it can be used in calculations
>>> x = np.array([(1, 2),(3,4)], dtype=[('a', np.int8), ('b', np.int8)]) >>> xv = x.view(dtype=np.int8).reshape(-1,2) >>> xv array([[1, 2], [3, 4]], dtype=int8) >>> xv.mean(0) array([2., 3.])
Making changes to the view changes the underlying array
>>> xv[0,1] = 20 >>> x array([(1, 20), (3, 4)], dtype=[('a', 'i1'), ('b', 'i1')])
Using a view to convert an array to a recarray:
>>> z = x.view(np.recarray) >>> z.a array([1, 3], dtype=int8)
Views share data:
>>> x[0] = (9, 10) >>> z[0] (9, 10)
Views that change the dtype size (bytes per entry) should normally be avoided on arrays defined by slices, transposes, fortran-ordering, etc.:
>>> x = np.array([[1,2,3],[4,5,6]], dtype=np.int16) >>> y = x[:, 0:2] >>> y array([[1, 2], [4, 5]], dtype=int16) >>> y.view(dtype=[('width', np.int16), ('length', np.int16)]) Traceback (most recent call last): ... ValueError: To change to a dtype of a different size, the array must be C-contiguous >>> z = y.copy() >>> z.view(dtype=[('width', np.int16), ('length', np.int16)]) array([[(1, 2)], [(4, 5)]], dtype=[('width', '<i2'), ('length', '<i2')])
-
write(self, target, *args, **kwargs)[source]¶ Write this
Spectrogramto a fileArguments and keywords depend on the output format, see the online documentation for full details for each format, the parameters below are common to most formats.
- Parameters
target :
stroutput filename
format :
str, optionaloutput format identifier. If not given, the format will be detected if possible. See below for list of acceptable formats.
Notes
The available built-in formats are:
Format
Read
Write
Auto-identify
hdf5
Yes
Yes
No
-
zip(self)[source]¶ Zip the
xindexandvaluearrays of thisSeries- Returns
stacked : 2-d
numpy.ndarray
Examples
>>> a = Series([0, 2, 4, 6, 8], xindex=[-5, -4, -3, -2, -1]) >>> a.zip() array([[-5., 0.], [-4., 2.], [-3., 4.], [-2., 6.], [-1., 8.]])
-
zpk(self, zeros, poles, gain, analog=True)[source]¶ Filter this
Spectrogramby applying a zero-pole-gain filter- Parameters
zeros :
array-likelist of zero frequencies (in Hertz)
poles :
array-likelist of pole frequencies (in Hertz)
gain :
floatDC gain of filter
analog :
bool, optionaltype of ZPK being applied, if
analog=Trueall parameters will be converted in the Z-domain for digital filtering- Returns
specgram :
Spectrogramthe frequency-domain filtered version of the input data
See also
Spectrogram.filterfor details on how a digital ZPK-format filter is applied
Examples
To apply a zpk filter with file poles at 100 Hz, and five zeros at 1 Hz (giving an overall DC gain of 1e-10):
>>> data2 = data.zpk([100]*5, [1]*5, 1e-10)
-
property
band Frequency band described by this
Spectrogram
-
crop_frequencies(self, low=None, high=None, copy=False)[source] Crop this
Spectrogramto the specified frequencies- Parameters
low :
floatlower frequency bound for cropped
Spectrogramhigh :
floatupper frequency bound for cropped
Spectrogramcopy :
boolif
Falsereturn a view of the original data, otherwise create a fresh memory copy- Returns
spec :
SpectrogramA new
Spectrogramwith a subset of data from the frequency axis
-
property
df Frequency spacing of this
Spectrogram- Type
Quantityin Hertz
-
property
dt Time-spacing for this
Spectrogram- Type
Quantityin seconds
-
property
epoch Starting GPS epoch for this
Spectrogram- Type
-
property
f0 Starting frequency for this
Spectrogram- Type
Quantityin Hertz
-
filter(self, *filt, **kwargs)[source] Apply the given filter to this
Spectrogram.- Parameters
*filt : filter arguments
analog :
bool, optionalinplace :
bool, optional- Returns
result :
Spectrogramthe filtered version of the input
Spectrogram, ifinplace=Truewas given, this is just a reference to the modified input array- Raises
ValueError
if
filtarguments cannot be interpreted properly
-
property
frequencies Series of frequencies for this Spectrogram
-
classmethod
from_spectra(*spectra, **kwargs)[source] Build a new
Spectrogramfrom a list of spectra.- Parameters
*spectra
any number of
FrequencySeriesseriesdt :
float,Quantity, optionalstride between given spectra
- Returns
Spectrogram
a new
Spectrogramfrom a vertical stacking of the spectra The new object takes the metadata from the first givenFrequencySeriesif not given explicitly
Notes
Each
FrequencySeriespassed to this constructor must be the same length.
-
percentile(self, percentile)[source] Calculate a given spectral percentile for this
Spectrogram.- Parameters
percentile :
floatpercentile (0 - 100) of the bins to compute
- Returns
spectrum :
FrequencySeriesthe given percentile
FrequencySeriescalculated from thisSpectralVaraicence
-
plot(self, figsize=(12, 6), xscale='auto-gps', **kwargs)[source] Plot the data for this
Spectrogram- Parameters
**kwargs
all keyword arguments are passed along to underlying functions, see below for references
- Returns
plot :
Plotthe
Plotcontaining the data
See also
matplotlib.pyplot.figurefor documentation of keyword arguments used to create the figure
matplotlib.figure.Figure.add_subplotfor documentation of keyword arguments used to create the axes
gwpy.plot.Axes.imshowgwpy.plot.Axes.pcolormeshfor documentation of keyword arguments used in rendering the
Spectrogramdata
-
ratio(self, operand)[source] Calculate the ratio of this
Spectrogramagainst a reference- Parameters
operand :
str,FrequencySeries,Quantitya
FrequencySeriesorQuantityto weight against, or one of'mean': weight against the mean of each spectrum in this Spectrogram'median': weight against the median of each spectrum in this Spectrogram
- Returns
spectrogram :
Spectrograma new
Spectrogram- Raises
ValueError
if
operandis given as astrthat isn’t supported
-
classmethod
read(source, *args, **kwargs)[source] Read data into a
SpectrogramArguments and keywords depend on the output format, see the online documentation for full details for each format, the parameters below are common to most formats.
- Parameters
-
*args
Other arguments are (in general) specific to the given
format.format :
str, optionalSource format identifier. If not given, the format will be detected if possible. See below for list of acceptable formats.
**kwargs
Other keywords are (in general) specific to the given
format. - Returns
- specgram
Spectrogram
- specgram
Notes
The available built-in formats are:
Format
Read
Write
Auto-identify
hdf5
Yes
No
No
-
property
span GPS [start, stop) span for this
Spectrogram- Type
-
property
t0 GPS time of first time bin
- Type
Quantityin seconds
-
property
times Series of GPS times for each sample
-
variance(self, bins=None, low=None, high=None, nbins=500, log=False, norm=False, density=False)[source] Calculate the
SpectralVarianceof thisSpectrogram.- Parameters
bins :
ndarray, optional, defaultNonearray of histogram bin edges, including the rightmost edge
low :
float, optional, default:Noneleft edge of lowest amplitude bin, only read if
binsis not givenhigh :
float, optional, default:Noneright edge of highest amplitude bin, only read if
binsis not givennbins :
int, optional, default:500number of bins to generate, only read if
binsis not givenlog :
bool, optional, default:Falsecalculate amplitude bins over a logarithmic scale, only read if
binsis not givennorm :
bool, optional, default:Falsenormalise bin counts to a unit sum
density :
bool, optional, default:Falsenormalise bin counts to a unit integral
- Returns
specvar :
SpectralVariance2D-array of spectral frequency-amplitude counts
See also
numpy.histogramfor details on specifying bins and weights
-
write(self, target, *args, **kwargs)[source] Write this
Spectrogramto a fileArguments and keywords depend on the output format, see the online documentation for full details for each format, the parameters below are common to most formats.
- Parameters
target :
stroutput filename
format :
str, optionaloutput format identifier. If not given, the format will be detected if possible. See below for list of acceptable formats.
Notes
The available built-in formats are:
Format
Read
Write
Auto-identify
hdf5
Yes
Yes
No
-
zpk(self, zeros, poles, gain, analog=True)[source] Filter this
Spectrogramby applying a zero-pole-gain filter- Parameters
zeros :
array-likelist of zero frequencies (in Hertz)
poles :
array-likelist of pole frequencies (in Hertz)
gain :
floatDC gain of filter
analog :
bool, optionaltype of ZPK being applied, if
analog=Trueall parameters will be converted in the Z-domain for digital filtering- Returns
specgram :
Spectrogramthe frequency-domain filtered version of the input data
See also
Spectrogram.filterfor details on how a digital ZPK-format filter is applied
Examples
To apply a zpk filter with file poles at 100 Hz, and five zeros at 1 Hz (giving an overall DC gain of 1e-10):
>>> data2 = data.zpk([100]*5, [1]*5, 1e-10)